Jump to navigation
Jump to search
NCBI: 03-AUG-2016
⊟Summary[edit | edit source]
- organism: Staphylococcus aureus NCTC8325
- locus tag: SAOUHSC_00928
- pan locus tag?: SAUPAN003146000
- symbol: SAOUHSC_00928
- pan gene symbol?: opp4A
- synonym:
- product: oligopeptide ABC transporter substrate-binding protein
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
- type: CDS
- locus tag: SAOUHSC_00928
- symbol: SAOUHSC_00928
- product: oligopeptide ABC transporter substrate-binding protein
- replicon: chromosome
- strand: +
- coordinates: 899923..901638
- length: 1716
- essential: no DEG other strains
⊟Accession numbers[edit | edit source]
- Gene ID: 3920757 NCBI
- RefSeq: YP_499481 NCBI
- BioCyc: G1I0R-870 BioCyc
- MicrobesOnline: 1289392 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421
481
541
601
661
721
781
841
901
961
1021
1081
1141
1201
1261
1321
1381
1441
1501
1561
1621
1681ATGGGGAAGCTAATTAAATATATTTCAATACTTCTTATTGTCGTTTTAGTGTTGAGTGCT
TGCGGAAAAAGCAGTAATAAAGATGAAGGAGTAAAAGATGCTACTAAAACGGAAACCTCA
AAACATAAAGGTGGTACCTTAAATGTAGCATTAACAGCACCGCCAAGTGGTGTTTATTCT
TCGTTATTAAATAGTACACATGCAGATTCTGTAGTTGAGGGATATTTTAACGAAAGCTTA
TTAGCAACTGATAAAAAAATACGTCCTAAGGCATATATTGCTTCATGGAAGGACATCGAG
CCGGCTAAGAAAATAGAATTTAAAATTAAAAAAGGTATTAAATGGCATGATGGTAATGAA
TTGAAAATTGATGATTGGATTTATTCAATTGAAGTCTTAGCTAACAAGGACTACGAAGGT
GCTTATTATCCAAGTGTAGAAAATATCCAAGGTGCGAAAGATTATCATGAAGGAAAAACT
GATCATATTAGCGGATTGAAGAAAATAGATGACTACACTATGCAGGTTACATTTGATAAA
AAACAAGAAAATTACTTAACAGGATTTATTACTGGACCTTTATTAAGTAAAAAATATTTA
TCAGATGTACCAATTAAAGATTTAGCGAAATCAGATAAAATCCGAAAATATCCTATTGGT
ATTGGACCGTATAAAGTTAAGAAAATCGTTCCAGGTGAGGCTGTTCAACTCGTTAAATTT
GATGATTATTGGCAAGGTAAGCCTGCACTAGACAAAATCAATTTAAAAGTTATTGATCAA
GCGCAAATTATTAAGGCAATGGAAAAAGGCGATATTGATGTTGCGAATGATGCTACCGGT
GCAATGGCAAAAGATGCTAAGTCATCTAATGCTGGTCTCAAGGTATTATCTGCGCCAAGC
TTAGACTACGGTTTAATAGGATTCGTATCTCATGATTACGATAAAAAAGCTAATAAAACT
GGTAAAGTGAGACCAAAATATGAAGACAAAGAATTACGTAAAGCAATGCTTTATGCAATT
GATAGAGAAAAATGGATCAAAGCGTTTTTCAATGGTTACGCTAGTGAAATCAATAGTTTT
GTACCATCTATGCATTGGATAGCAGCCAATCCTAAGGACCTAAATGATTACAAATATGAT
CCTGAAAAAGCTAAAAAAATCTTAGATAAGTTAGGTTATAAAGATAGAGATGGTGACGGA
TTTAGAGAAGATCCTAAAGGTAATAAATTTGAGATTAACTTTAAACATAATTCAGGTTCT
AATCCTACTTTTGAACCAAGAACTGCTGCGATAAAAGATTTCTGGGAAAAAGTTGGCTTG
AAAACAAATGTGAAGTTAGTAGAATTCGGTAAATATAATGAAGACTTAGCAAATGCATCT
AAAGATATGGAAGTGTACTTCAGATCATGGGCAGGAGGTACAGATCCAGATCCATCAGAT
TTATACCACACTGATAGACCTCAAAATGAAATGAGAACAGTTTTACCAAAATCAGATCAA
TATTTAGATGATGCATTAGACTTCGAAAAAGTAGGCATTGATGAAAAGAAACGTAAAGAT
ATTTATGTTAAATGGCAAAAATATATGAATGATGAGTTACCTGGATTACCAATGTTCCAA
GGTAAATCGATAACTATTGTTAACGATAAAGTACGAAACTTAGACATTGAAATTGGAACT
GATCAAAGTTTATATAATTTAACTAAAGAAGCTTAG60
120
180
240
300
360
420
480
540
600
660
720
780
840
900
960
1020
1080
1140
1200
1260
1320
1380
1440
1500
1560
1620
1680
1716
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SAOUHSC_00928
- symbol: SAOUHSC_00928
- description: oligopeptide ABC transporter substrate-binding protein
- length: 571
- theoretical pI: 9.01283
- theoretical MW: 64527
- GRAVY: -0.640455
⊟Function[edit | edit source]
- TIGRFAM: Transport and binding proteins Cations and iron carrying compounds nickel ABC transporter, nickel/metallophore periplasmic binding protein (TIGR02294; HMM-score: 93)Transport and binding proteins Unknown substrate ABC transporter substrate binding protein, KPN_01854 family (TIGR04028; HMM-score: 79.9)
- TheSEED :
- Oligopeptide ABC transporter, periplasmic oligopeptide-binding protein OppA (TC 3.A.1.5.1)
Membrane Transport ABC transporters ABC transporter oligopeptide (TC 3.A.1.5.1) Oligopeptide ABC transporter, periplasmic oligopeptide-binding protein OppA (TC 3.A.1.5.1)and 1 more - PFAM: PBP (CL0177) SBP_bac_5; Bacterial extracellular solute-binding proteins, family 5 Middle (PF00496; HMM-score: 265.4)and 1 moreGBD (CL0202) P_proprotein; Proprotein convertase P-domain (PF01483; HMM-score: 12.5)
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors:
- effectors:
⊟Localization[edit | edit source]
- PSORTb: Cellwall
- Cytoplasmic Score: 0
- Cytoplasmic Membrane Score: 0.53
- Cellwall Score: 8.76
- Extracellular Score: 0.7
- Internal Helices: 0
- LocateP: Lipid anchored
- Prediction by SwissProt Classification: Extracellular
- Pathway Prediction: Sec-(SPII)
- Intracellular possibility: 0
- Signal peptide possibility: 0.5
- N-terminally Anchored Score: 1
- Predicted Cleavage Site: LVLSACG
- SignalP: Signal peptide LIPO(Sec/SPII) length 20 aa
- SP(Sec/SPI): 0.000481
- TAT(Tat/SPI): 0.000083
- LIPO(Sec/SPII): 0.99921
- Cleavage Site: CS pos: 20-21. LSA-CG. Pr: 0.9997
- predicted transmembrane helices (TMHMM): 0
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MGKLIKYISILLIVVLVLSACGKSSNKDEGVKDATKTETSKHKGGTLNVALTAPPSGVYSSLLNSTHADSVVEGYFNESLLATDKKIRPKAYIASWKDIEPAKKIEFKIKKGIKWHDGNELKIDDWIYSIEVLANKDYEGAYYPSVENIQGAKDYHEGKTDHISGLKKIDDYTMQVTFDKKQENYLTGFITGPLLSKKYLSDVPIKDLAKSDKIRKYPIGIGPYKVKKIVPGEAVQLVKFDDYWQGKPALDKINLKVIDQAQIIKAMEKGDIDVANDATGAMAKDAKSSNAGLKVLSAPSLDYGLIGFVSHDYDKKANKTGKVRPKYEDKELRKAMLYAIDREKWIKAFFNGYASEINSFVPSMHWIAANPKDLNDYKYDPEKAKKILDKLGYKDRDGDGFREDPKGNKFEINFKHNSGSNPTFEPRTAAIKDFWEKVGLKTNVKLVEFGKYNEDLANASKDMEVYFRSWAGGTDPDPSDLYHTDRPQNEMRTVLPKSDQYLDDALDFEKVGIDEKKRKDIYVKWQKYMNDELPGLPMFQGKSITIVNDKVRNLDIEIGTDQSLYNLTKEA
⊟Experimental data[edit | edit source]
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
⊟Regulation[edit | edit source]
- regulator: CodY* (repression) regulon
CodY* (TF) important in Amino acid metabolism; RegPrecise
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: [1]
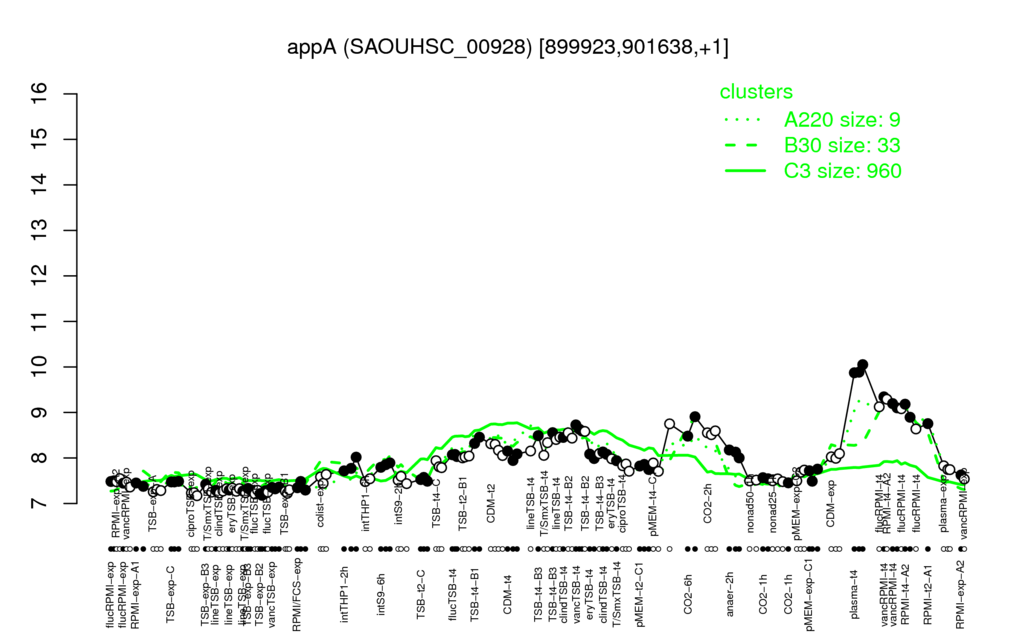 Multi-gene expression profiles
Multi-gene expression profiles
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
- Aureolib: no data available
⊟Protein stability[edit | edit source]
- half-life: no data available
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You are kindly invited to share additional interesting facts.
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ Ulrike Mäder, Pierre Nicolas, Maren Depke, Jan Pané-Farré, Michel Debarbouille, Magdalena M van der Kooi-Pol, Cyprien Guérin, Sandra Dérozier, Aurelia Hiron, Hanne Jarmer, Aurélie Leduc, Stephan Michalik, Ewoud Reilman, Marc Schaffer, Frank Schmidt, Philippe Bessières, Philippe Noirot, Michael Hecker, Tarek Msadek, Uwe Völker, Jan Maarten van Dijl
Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions.
PLoS Genet: 2016, 12(4);e1005962
[PubMed:27035918] [WorldCat.org] [DOI] (I e)
