⊟Summary[edit | edit source]
- organism: Staphylococcus aureus NCTC8325
- locus tag: S596 [1]
- symbol: IsrR
- synonym: S596
- product: sRNA
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
- type: misc_RNA
- locus tag: S596 [1]
- symbol: IsrR
- product: sRNA
- replicon: chromosome
- strand: +
- coordinates: 1362874..1363048
- length: 175
- essential: unknown
⊟Accession numbers[edit | edit source]
- SRD: srn_2975 SRD
⊟Phenotype[edit | edit source]
IsrR is required for optimum growth in iron-depleted media[2].
IsrR is required for full virulence in a mouse sepsis model of virulence[2].
IsrR downregulates nitrite production in anaerobic conditions[2].
IsrR associates by base-pairing to fdhA, gltB2, narG and nasD mRNAs to downregulate their translation[2].
IsrR downregulates aconitase (CitB) and its activitor CcpE[3].
IsrR associates by base-pairing to citB and ccpE mRNAs to downregulate their translation[3].
IsrR downregulates methylthiotransferase MiaB by preventing miaB mRNA translation[4].
- Share your knowledge and add information here. [edit]
- Functional name (symbol): isrR for iron sparing response RNA[2].
- IsrR and its regulation by Fur are conserved in the genus Staphylococcus[2].
- The isrR gene has two Fur boxes and is expressed under low iron growth conditions[2].
- IsrR has 3 C-rich regions (CRR) involved in pairing with its mRNA targets. Many targets and putative targets of IsrR are mRNAs expressing iron-containing enzymes[2].
- The putative function of IsrR is to downregulate non-essential iron-containing enzymes in order to spare iron in bacteria[2].
⊟DNA sequence[edit | edit source]
- 1
61
121TCACTAATGTATAATAGTAGTTGAAAATGATTATCAATACCACATAGAACATCCCCCCCA
CAACGTTTCGTTCTTGTTGGATTGGTCATTTTCAAATATTCCCCTTTTATATGCCCGTAA
AAGACAATATACGTTATAACAACGTTTTATAAAAGCAGTAAACCCTTACGACACT60
120
175
⊟Expression & Regulation[edit | edit source]
⊟Regulation[edit | edit source]
- regulator: Fur* (repression) regulon
Fur (repression)
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: [1]
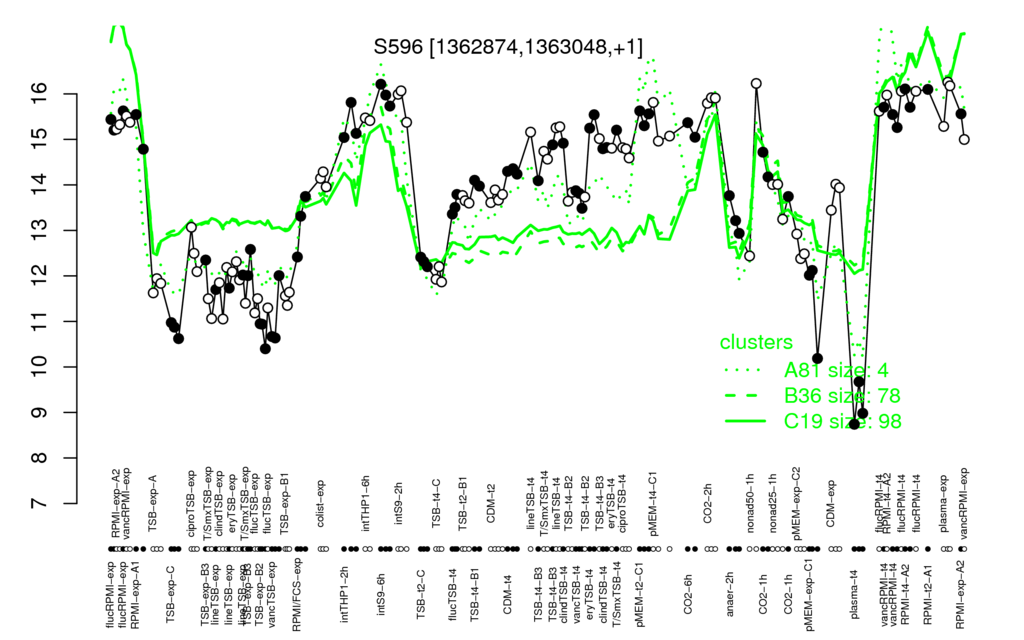 Multi-gene expression profiles
Multi-gene expression profiles
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You can add further information about the gene and protein here. [edit]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ 1.0 1.1 1.2 1.3 1.4 Ulrike Mäder, Pierre Nicolas, Maren Depke, Jan Pané-Farré, Michel Debarbouille, Magdalena M van der Kooi-Pol, Cyprien Guérin, Sandra Dérozier, Aurelia Hiron, Hanne Jarmer, Aurélie Leduc, Stephan Michalik, Ewoud Reilman, Marc Schaffer, Frank Schmidt, Philippe Bessières, Philippe Noirot, Michael Hecker, Tarek Msadek, Uwe Völker, Jan Maarten van Dijl
Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions.
PLoS Genet: 2016, 12(4);e1005962
[PubMed:27035918] [WorldCat.org] [DOI] (I e) - ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 2.7 2.8 Rodrigo H Coronel-Tellez, Mateusz Pospiech, Maxime Barrault, Wenfeng Liu, Valérie Bordeau, Christelle Vasnier, Brice Felden, Bruno Sargueil, Philippe Bouloc
sRNA-controlled iron sparing response in Staphylococci.
Nucleic Acids Res: 2022, 50(15);8529-8546
[PubMed:35904807] [WorldCat.org] [DOI] (I p) - ↑ 3.0 3.1 Maxime Barrault, Svetlana Chabelskaya, Rodrigo H Coronel-Tellez, Claire Toffano-Nioche, Eric Jacquet, Philippe Bouloc
Staphylococcal aconitase expression during iron deficiency is controlled by an sRNA-driven feedforward loop and moonlighting activity.
Nucleic Acids Res: 2024;
[PubMed:38869061] [WorldCat.org] [DOI] (I a) - ↑ Maxime Barrault, Elise Leclair, Etornam Kofi Kumeko, Eric Jacquet, Philippe Bouloc
Staphylococcal sRNA IsrR downregulates methylthiotransferase MiaB under iron-deficient conditions.
Microbiol Spectr: 2024;e0388823
[PubMed:39162503] [WorldCat.org] [DOI] (I a) - ↑ Rodrigo H Coronel-Tellez, Mateusz Pospiech, Maxime Barrault, Wenfeng Liu, Valérie Bordeau, Christelle Vasnier, Brice Felden, Bruno Sargueil, Philippe Bouloc
sRNA-controlled iron sparing response in Staphylococci.
Nucleic Acids Res: 2022, 50(15);8529-8546
[PubMed:35904807] [WorldCat.org] [DOI] (I p)
⊟Relevant publications[edit | edit source]
Rodrigo H Coronel-Tellez, Mateusz Pospiech, Maxime Barrault, Wenfeng Liu, Valérie Bordeau, Christelle Vasnier, Brice Felden, Bruno Sargueil, Philippe Bouloc
sRNA-controlled iron sparing response in Staphylococci.
Nucleic Acids Res: 2022, 50(15);8529-8546
[PubMed:35904807] [WorldCat.org] [DOI] (I p)Alexander Ganske, Larissa Milena Busch, Christian Hentschker, Alexander Reder, Stephan Michalik, Kristin Surmann, Uwe Völker, Ulrike Mäder
Exploring the targetome of IsrR, an iron-regulated sRNA controlling the synthesis of iron-containing proteins in Staphylococcus aureus.
Front Microbiol: 2024, 15;1439352
[PubMed:39035440] [WorldCat.org] [DOI] (P e)
