Jump to navigation
Jump to search
NCBI: 03-AUG-2016
⊟Summary[edit | edit source]
- organism: Staphylococcus aureus NCTC8325
- locus tag: SAOUHSC_00005
- pan locus tag?: SAUPAN000011000
- symbol: SAOUHSC_00005
- pan gene symbol?: gyrB
- synonym:
- product: DNA gyrase subunit B
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
⊟Accession numbers[edit | edit source]
- Gene ID: 3919178 NCBI
- RefSeq: YP_498613 NCBI
- BioCyc: G1I0R-5 BioCyc
- MicrobesOnline: 1288505 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421
481
541
601
661
721
781
841
901
961
1021
1081
1141
1201
1261
1321
1381
1441
1501
1561
1621
1681
1741
1801
1861
1921ATGGTGACTGCATTGTCAGATGTAAACAACACGGATAATTATGGTGCTGGGCAAATACAA
GTATTAGAAGGTTTAGAAGCAGTACGTAAAAGACCAGGTATGTATATAGGATCGACTTCA
GAGAGAGGTTTGCACCATTTAGTGTGGGAAATTGTCGATAATAGTATCGATGAAGCATTA
GCTGGTTATGCAAATCAAATTGAAGTTGTTATTGAAAAAGATAACTGGATTAAAGTAACG
GATAACGGACGTGGTATCCCAGTTGATATTCAAGAAAAAATGGGACGTCCAGCTGTCGAA
GTTATTTTAACTGTTTTACATGCTGGTGGTAAATTTGGCGGTGGCGGATACAAAGTATCT
GGTGGTTTACATGGTGTTGGTTCATCAGTTGTAAACGCATTGTCACAAGACTTAGAAGTA
TATGTACACAGAAATGAGACTATATATCATCAAGCATATAAAAAAGGTGTACCTCAATTT
GACTTAAAAGAAGTTGGCACAACTGATAAGACAGGTACTGTCATTCGTTTTAAAGCAGAT
GGAGAAATCTTCACAGAGACAACTGTATACAACTATGAAACATTACAGCAGCGTATTAGA
GAGCTTGCTTTCTTAAACAAAGGAATTCAAATCACATTAAGAGATGAACGTGATGAAGAA
AACGTTAGAGAAGACTCCTATCACTATGAGGGCGGTATTAAATCGTACGTTGAGTTATTG
AACGAAAATAAAGAACCTATTCATGATGAGCCAATTTATATTCATCAATCTAAAGATGAT
ATTGAAGTAGAAATTGCGATTCAATATAACTCAGGATATGCCACAAATCTTTTAACTTAC
GCAAATAACATTCATACGTATGAAGGTGGTACGCATGAAGACGGATTCAAACGTGCATTA
ACGCGTGTCTTAAATAGTTATGGTTTAAGTAGCAAGATTATGAAAGAAGAAAAAGATAGA
CTTTCTGGTGAAGATACACGTGAAGGTATGACAGCAATTATATCTATCAAACATGGTGAT
CCTCAATTCGAAGGTCAAACGAAGACAAAATTAGGTAATTCTGAAGTGCGTCAAGTTGTA
GATAAATTATTCTCAGAGCACTTTGAACGATTTTTATATGAAAATCCACAAGTCGCACGT
ACAGTGGTTGAAAAAGGTATTATGGCGGCACGTGCACGTGTTGCTGCGAAAAAAGCGCGT
GAAGTAACACGTCGTAAATCAGCGTTAGATGTAGCAAGCCTTCCAGGTAAATTAGCCGAT
TGCTCTAGTAAAAGTCCTGAAGAATGTGAGATTTTCTTAGTCGAAGGGGACTCTGCCGGG
GGGTCTACAAAATCTGGTCGTGACTCTAGAACGCAGGCGATTTTACCATTACGAGGTAAG
ATATTAAATGTTGAAAAAGCACGATTAGATAGAATTTTGAATAACAATGAAATTCGTCAA
ATGATCACAGCATTTGGTACAGGAATCGGTGGCGACTTTGATCTAGCGAAAGCAAGATAT
CACAAAATCGTCATTATGACTGATGCCGATGTGGATGGAGCGCATATTAGAACATTGTTA
TTAACATTCTTCTATCGATTTATGAGACCGTTAATTGAAGCAGGCTATGTGTATATTGCA
CAGCCACCGTTGTATAAACTGACACAAGGTAAACAAAAGTATTATGTATACAATGATAGG
GAACTTGATAAACTTAAATCTGAATTGAATCCAACACCAAAATGGTCTATTGCACGATAC
AAAGGTCTTGGAGAAATGAATGCAGATCAATTATGGGAAACAACAATGAACCCTGAGCAC
CGCGCTCTTTTACAAGTAAAACTTGAAGATGCGATTGAAGCGGACCAAACATTTGAAATG
TTAATGGGTGACGTTGTAGAAAACCGTAGACAATTTATAGAAGATAATGCAGTTTATGCA
AACTTAGACTTCTAA60
120
180
240
300
360
420
480
540
600
660
720
780
840
900
960
1020
1080
1140
1200
1260
1320
1380
1440
1500
1560
1620
1680
1740
1800
1860
1920
1935
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SAOUHSC_00005
- symbol: SAOUHSC_00005
- description: DNA gyrase subunit B
- length: 644
- theoretical pI: 5.68145
- theoretical MW: 72539.3
- GRAVY: -0.489907
⊟Function[edit | edit source]
- reaction: EC 5.99.1.3? ExPASyDNA topoisomerase (ATP-hydrolyzing) ATP-dependent breakage, passage and rejoining of double-stranded DNA
- TIGRFAM: DNA metabolism DNA replication, recombination, and repair DNA gyrase, B subunit (TIGR01059; EC 5.99.1.3; HMM-score: 994)DNA metabolism DNA replication, recombination, and repair DNA topoisomerase IV, B subunit (TIGR01058; EC 5.99.1.-; HMM-score: 828)and 3 moreDNA metabolism DNA replication, recombination, and repair DNA topoisomerase IV, B subunit (TIGR01055; EC 5.99.1.-; HMM-score: 605.8)DNA metabolism DNA replication, recombination, and repair DNA mismatch repair protein MutL (TIGR00585; HMM-score: 20.1)DNA metabolism DNA replication, recombination, and repair DNA topoisomerase VI, B subunit (TIGR01052; EC 5.99.1.3; HMM-score: 16.2)
- TheSEED :
- DNA gyrase subunit B (EC 5.99.1.3)
DNA Metabolism DNA replication DNA topoisomerases, Type II, ATP-dependent DNA gyrase subunit B (EC 5.99.1.3)and 1 more - PFAM: S5 (CL0329) DNA_gyraseB; DNA gyrase B (PF00204; HMM-score: 211.5)and 6 moreno clan defined DNA_gyraseB_C; DNA gyrase B subunit, carboxyl terminus (PF00986; HMM-score: 106.8)Toprim-like (CL0413) Toprim; Toprim domain (PF01751; HMM-score: 66.5)His_Kinase_A (CL0025) HATPase_c; Histidine kinase-, DNA gyrase B-, and HSP90-like ATPase (PF02518; HMM-score: 62.7)no clan defined TOPRIM_C; C-terminal associated domain of TOPRIM (PF16898; HMM-score: 17.2)His_Kinase_A (CL0025) HATPase_c_3; Histidine kinase-, DNA gyrase B-, and HSP90-like ATPase (PF13589; HMM-score: 15.9)no clan defined DUF2110_N; DUF2110 N-terminal domain (PF24872; HMM-score: 14)
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors: Mg2+, Mn2+
- effectors:
⊟Localization[edit | edit source]
- PSORTb: Cytoplasmic
- Cytoplasmic Score: 9.97
- Cytoplasmic Membrane Score: 0
- Cellwall Score: 0.01
- Extracellular Score: 0.02
- Internal Helices: 0
- DeepLocPro: Cytoplasmic
- Cytoplasmic Score: 0.9534
- Cytoplasmic Membrane Score: 0.0052
- Cell wall & surface Score: 0.0002
- Extracellular Score: 0.0412
- LocateP: Intracellular
- Prediction by SwissProt Classification: Cytoplasmic
- Pathway Prediction: No pathway
- Intracellular possibility: 1
- Signal peptide possibility: -1
- N-terminally Anchored Score: 1
- Predicted Cleavage Site: No CleavageSite
- SignalP: no predicted signal peptide
- SP(Sec/SPI): 0.002817
- TAT(Tat/SPI): 0.000288
- LIPO(Sec/SPII): 0.000389
- predicted transmembrane helices (TMHMM): 0
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MVTALSDVNNTDNYGAGQIQVLEGLEAVRKRPGMYIGSTSERGLHHLVWEIVDNSIDEALAGYANQIEVVIEKDNWIKVTDNGRGIPVDIQEKMGRPAVEVILTVLHAGGKFGGGGYKVSGGLHGVGSSVVNALSQDLEVYVHRNETIYHQAYKKGVPQFDLKEVGTTDKTGTVIRFKADGEIFTETTVYNYETLQQRIRELAFLNKGIQITLRDERDEENVREDSYHYEGGIKSYVELLNENKEPIHDEPIYIHQSKDDIEVEIAIQYNSGYATNLLTYANNIHTYEGGTHEDGFKRALTRVLNSYGLSSKIMKEEKDRLSGEDTREGMTAIISIKHGDPQFEGQTKTKLGNSEVRQVVDKLFSEHFERFLYENPQVARTVVEKGIMAARARVAAKKAREVTRRKSALDVASLPGKLADCSSKSPEECEIFLVEGDSAGGSTKSGRDSRTQAILPLRGKILNVEKARLDRILNNNEIRQMITAFGTGIGGDFDLAKARYHKIVIMTDADVDGAHIRTLLLTFFYRFMRPLIEAGYVYIAQPPLYKLTQGKQKYYVYNDRELDKLKSELNPTPKWSIARYKGLGEMNADQLWETTMNPEHRALLQVKLEDAIEADQTFEMLMGDVVENRRQFIEDNAVYANLDF
⊟Experimental data[edit | edit source]
- experimentally validated: PeptideAtlas [2] [3]
- protein localization: data available for COL
- quantitative data / protein copy number per cell: data available for COL
- interaction partners:
SAOUHSC_01246 (infB) translation initiation factor IF-2 [4] (data from MRSA252) SAOUHSC_00006 DNA gyrase subunit A [4] (data from MRSA252) SAOUHSC_00529 elongation factor G [4] (data from MRSA252) SAOUHSC_01806 pyruvate kinase [4] (data from MRSA252)
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
⊟Regulation[edit | edit source]
- regulator:
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: [5]
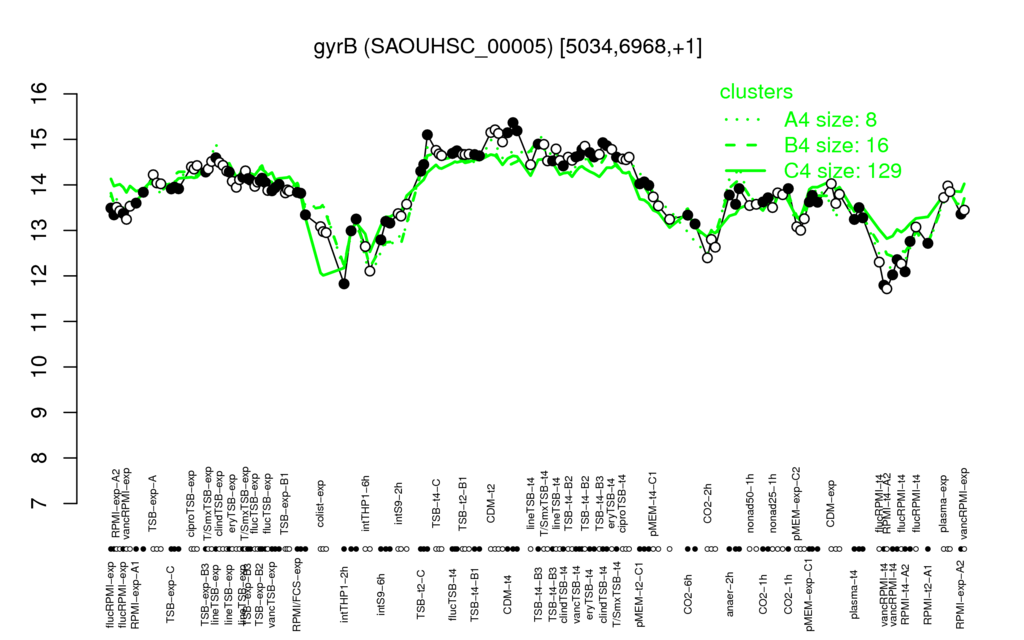 Multi-gene expression profiles
Multi-gene expression profiles
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
⊟Protein stability[edit | edit source]
- half-life: no data available
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You can add further information about the gene and protein here. [edit]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ Roy R Chaudhuri, Andrew G Allen, Paul J Owen, Gil Shalom, Karl Stone, Marcus Harrison, Timothy A Burgis, Michael Lockyer, Jorge Garcia-Lara, Simon J Foster, Stephen J Pleasance, Sarah E Peters, Duncan J Maskell, Ian G Charles
Comprehensive identification of essential Staphylococcus aureus genes using Transposon-Mediated Differential Hybridisation (TMDH).
BMC Genomics: 2009, 10;291
[PubMed:19570206] [WorldCat.org] [DOI] (I e) - ↑ Maren Depke, Stephan Michalik, Alexander Rabe, Kristin Surmann, Lars Brinkmann, Nico Jehmlich, Jörg Bernhardt, Michael Hecker, Bernd Wollscheid, Zhi Sun, Robert L Moritz, Uwe Völker, Frank Schmidt
A peptide resource for the analysis of Staphylococcus aureus in host-pathogen interaction studies.
Proteomics: 2015, 15(21);3648-61
[PubMed:26224020] [WorldCat.org] [DOI] (I p) - ↑ Stephan Michalik, Maren Depke, Annette Murr, Manuela Gesell Salazar, Ulrike Kusebauch, Zhi Sun, Tanja C Meyer, Kristin Surmann, Henrike Pförtner, Petra Hildebrandt, Stefan Weiss, Laura Marcela Palma Medina, Melanie Gutjahr, Elke Hammer, Dörte Becher, Thomas Pribyl, Sven Hammerschmidt, Eric W Deutsch, Samuel L Bader, Michael Hecker, Robert L Moritz, Ulrike Mäder, Uwe Völker, Frank Schmidt
A global Staphylococcus aureus proteome resource applied to the in vivo characterization of host-pathogen interactions.
Sci Rep: 2017, 7(1);9718
[PubMed:28887440] [WorldCat.org] [DOI] (I e) - ↑ 4.0 4.1 4.2 4.3 Artem Cherkasov, Michael Hsing, Roya Zoraghi, Leonard J Foster, Raymond H See, Nikolay Stoynov, Jihong Jiang, Sukhbir Kaur, Tian Lian, Linda Jackson, Huansheng Gong, Rick Swayze, Emily Amandoron, Farhad Hormozdiari, Phuong Dao, Cenk Sahinalp, Osvaldo Santos-Filho, Peter Axerio-Cilies, Kendall Byler, William R McMaster, Robert C Brunham, B Brett Finlay, Neil E Reiner
Mapping the protein interaction network in methicillin-resistant Staphylococcus aureus.
J Proteome Res: 2011, 10(3);1139-50
[PubMed:21166474] [WorldCat.org] [DOI] (I p) - ↑ 5.0 5.1 Ulrike Mäder, Pierre Nicolas, Maren Depke, Jan Pané-Farré, Michel Debarbouille, Magdalena M van der Kooi-Pol, Cyprien Guérin, Sandra Dérozier, Aurelia Hiron, Hanne Jarmer, Aurélie Leduc, Stephan Michalik, Ewoud Reilman, Marc Schaffer, Frank Schmidt, Philippe Bessières, Philippe Noirot, Michael Hecker, Tarek Msadek, Uwe Völker, Jan Maarten van Dijl
Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions.
PLoS Genet: 2016, 12(4);e1005962
[PubMed:27035918] [WorldCat.org] [DOI] (I e)
