Jump to navigation
Jump to search
NCBI: 03-AUG-2016
⊟Summary[edit | edit source]
- organism: Staphylococcus aureus NCTC8325
- locus tag: SAOUHSC_00144
- pan locus tag?: SAUPAN001018000
- symbol: SAOUHSC_00144
- pan gene symbol?: ausA
- synonym:
- product: hypothetical protein
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
- type: CDS
- locus tag: SAOUHSC_00144
- symbol: SAOUHSC_00144
- product: hypothetical protein
- replicon: chromosome
- strand: +
- coordinates: 149045..156220
- length: 7176
- essential: no DEG other strains
⊟Accession numbers[edit | edit source]
- Gene ID: 3919852 NCBI
- RefSeq: YP_498743 NCBI
- BioCyc: G1I0R-134 BioCyc
- MicrobesOnline: 1288637 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421
481
541
601
661
721
781
841
901
961
1021
1081
1141
1201
1261
1321
1381
1441
1501
1561
1621
1681
1741
1801
1861
1921
1981
2041
2101
2161
2221
2281
2341
2401
2461
2521
2581
2641
2701
2761
2821
2881
2941
3001
3061
3121
3181
3241
3301
3361
3421
3481
3541
3601
3661
3721
3781
3841
3901
3961
4021
4081
4141
4201
4261
4321
4381
4441
4501
4561
4621
4681
4741
4801
4861
4921
4981
5041
5101
5161
5221
5281
5341
5401
5461
5521
5581
5641
5701
5761
5821
5881
5941
6001
6061
6121
6181
6241
6301
6361
6421
6481
6541
6601
6661
6721
6781
6841
6901
6961
7021
7081
7141ATGATTATGGGTAATTTGAGATTTCAACAGGAATATTTTCGTATATACAAAAATAATACA
GAATCAACGACACACCGTAATGCGTATTGGGTTAAACTCGCTAAAAATGTTGAAGCTACT
AAAATGATGTATGCATTATCGACAATTGTGCAACAACATGCATCTATAAGACATTTTTTT
GATGTTACTACCGATGACAATTTAACAATGATACTTCATGAATTTCTGCCTTTTATTGAG
ATAAAACAAGTTCCATCTTCTTCCGCAAACTATGATTTAGAAGCTTTTTTTAAGCAAGAA
TTAAGTACTTACCATTTTAATGATTCACCTTTATTCAAAGTTAAATTGTTTCAGTTCGCT
GATGCTGCATATATACTATTAGATTTTCATGTGTCCATTTTCGATGATAGTCAAATTGAT
ATTTTTCTTGATGATTTATGCAATGCATATCGTGGCAATACTGTTATTAACAATACTCGA
CAGCATGCACATATAAATAGAAATGATGATAAAGACAATCAAGATGCATCGCATATAGCA
TTAGACTCAAACTATTTTCGGTTAGAGAATAACTCTGACATCCATATTGATAGTTATTTT
CCAATTAAGCATCCATTTGAACAAGCTTTATATCAAACGTATTTGATTGATGATATGACA
TCAATAGATATGGCATCGTTGGCTGTTAGTGTGTATTTAGCTAATCATATAATGAGTCAA
CAACATGATGTCACATTAGGTATACATGTACCATCACATTTACCAAATGATTTACACGGA
AATATTGTGCCGTTAACGTTAACAATCGATGCAAAAGATGTATGTCAACGTTTTACAACA
GATTTTAATAAATGTGTGTTGCAAAATATGTCGCAATTACAGTGCGCGAAGTCTTCGCTT
TCACTAGAGACTATTTTTCATTGTTATCATCATATGATGTCTTGTTGTAATGATGTTATT
GAGGATGTACATCAAATACATGATGCACATACATCTTTAGCGGATATTGAAATTTTTCCA
CATCAACACGGGTTCAAAATTATATATAACAGTGCAGCATATGATTTGCTCTCAATCGAG
ACGCTGAGTGACTTAGTTCGAAATATTTATTTGCAAATTACTGAAGAAAATGGAAATAAA
CGAACAACTGTAGATGAACTTAATTTGATGACAGAACGTGATATTCAATTATATGACGAT
ATCAATTTAAGTTTGCCTGAGATAGATGATGCGCAAACAGTTGTTACCTTATTTGAGCAA
CAAGTTGAAGCAACGCCGAATCATGTCGCTGTGCAATTTGACGGAGTGTTTATAACATAT
CAAACATTGAATGCACGCGCGAATGATTTAGCACACCGTTTGAGAAACCAGTATGGTGTT
GAACCTAATGATCGTGTCGCTGTCATAGCTGAAAAAAGTATTGAGATGATAATAGCGATG
ATAGGTGTGTTGAAAGCTGGTGGGGCTTACGTGCCAATTGATCCGAACTATCCAAGTGAT
CGTCAGGAGTACATTTTAAAAGATGTAACGCCTAAAGTTGTAATAACGTACCAAGCTTTA
TATGAAAATGGTAAACAAAATATTAATCACATTGATTTGAATAAGATAGCGTGGAAAAAT
ATTGATAATCTTTCTAAATGTAACACGTTAGAAGATCATGCTTATGTTATTTACACGTCG
GGGACAACTGGTAACCCTAAAGGGACACTAATTCCGCACCGAGGTATTGTTCGCTTGGTC
CATCAAAATCATTATGTACCATTAAATGAAGAGACGACGATTTTGTTATCAGGAACTATA
GCCTTTGATGCTGCAACATTTGAAATATATGGTGCATTGCTCAATGGTGGAAAGCTGATT
GTTGCTAAAAAAGAACAATTATTAAATCCAATAGCGGTAGAACAATTAATCAATGAAAAT
GACGTTAATACTATGTGGTTAACCTCCTCATTATTTAATCAGATTGCTAGTGAACGAATA
GAAGTATTGGTACCGTTAAAGTATTTATTAATTGGTGGAGAAGTATTGAATGCTAAGTGG
GTGGATTTGCTTAATCAAAAACCGAAGCATCCTCAAATTATTAATGGTTATGGACCAACT
GAAAATACAACATTTACAACGACGTATAATATACCTAACAAAGTTCCAAATCGTATTCCT
ATTGGTAAACCGATTCTGGGTACTCATGTTTATATCATGCAAGGCGAGCGTCGGTGTGGC
GTTGGTATTCCTGGAGAATTATGTACAAGTGGCTTTGGGTTAGCTGCAGGTTATTTAAAT
CAGCCAGAATTGACAGCAGATAAATTTATCAAAGATTCAAATATAAATCAGCTGATGTAT
AGAAGTGGTGATATCGTTCGTTTGTTACCCGATGGCAACATAGATTATTTATATCGAAAG
GACAAACAAGTTAAGATTCGAGGGTTTAGGATTGAGTTGTCAGAGGTTGAGCATGCGCTC
GAGCGTATACAAGGTATTAATAAAGCAGTTGTTATTGTTCAAAATCATGATCAAGATCAG
TATATCGTTGCTTATTATGAAGCGATGCATACATTATCACATAATAAGATTAAATCACAA
TTACGTATGACCTTACCGGAGTACATGATACCAGTTAATTTCATGCATATTGAGCAAATT
CCTATTACTATTAATGGGAAATTAGATAAGAAGGCATTGCCTATCATGGACTATGTCGAT
ACGGATGCCTATGTAGCACCGAGTACAGATACCGAACACTTGCTATGCCAAATTTTTGCA
GATATTTTACATGTGAATCAAGTAGGTATTCATGATAATTTCTTTGAATTAGGTGGCCAT
TCATTAAAAGCAACGTTAGTGGTGAATCGGATAGAGGCATCTACTGGGAAACGATTACAA
ATTGGTGATTTATTACAAAAGCCAACTGTATTTGAACTAGCACAAGCGATTGCTAAGGTT
CAAGAACAAAACTATGAAGTGATTCCAGAAACTATAGTTAAAGATGATTATGTGCTGAGC
TCTGCACAAAAGCGTATGTATTTATTATGGAAATCAAACCATAAAGATACGGTGTATAAC
GTACCTTTTTTATGGCGGTTATCATCAGAACTTAATGTAGCTCAATTGCGACAAGCAGTG
CAGCGTTTGATAGCGCGACATGAGATTTTACGAACACAATATATTGTTGTAGATGATGAG
GTTCGACAACGTATTGTGGCAGATGTTGCAGTTGACTTTGAAGAAGTTAACACGCATTTT
ACGGATGAACAAGAAATCATGCGCCAATTTGTAGCACCTTTTAATTTGGAAAAGCCAAGT
CAAATTAGAGTGAGATACATTAGAAGTCCCTTACATGCATACCTCTTTATAGATACGCAT
CATATCATTAATGACGGTATGAGTAATATACAATTAATGAATGATCTTAACGCACTTTAT
CAACATAAATTATTGTTACCACTTAAATTGCAATATAAAGACTATAGTGAGTGGATGTCG
CATCGTGATATGACGAAACATAGACAATATTGGTTATCTCAATTCAAAGATGAAGTACCT
ATTTTAAGCTTACCGACAGACTATGTTAGACCAAATATTAAAACGACAAATGGAGCAATG
ATGTCATTTACAATGAATCAACAAATGAGACAGCTACTTCAAAAGTATGTAGAAAAGCAT
CAAATTACTGATTTTATGTTCTTTATGAGTGTGGTCATGACGTTGTTAAGTAGATATGCT
CGAAAAGATGATGTTGTTGTCGGTAGTGTGATGAGTGCGCGTATGCATAAAGGCACGGAG
CAAATGCTAGGCATGTTTGCTAATACGTTGGTATATAGAGGGCAACCGTCACCTGATAAA
ATGTGGACACAGTTTTTACAAGAGGTTAAGGAAATGAGTTTGGAGGCATACGAGCATCAA
GAATACCCATTCGAATGTTTAGTAAATGACTTAGATCAATCACATGATGCCTCACGGAAT
CCATTATTTGATGTCATGTTAGTACTACAAAACAATGAAACGAATCATGCTCATTTTGGG
CATAGTAAATTAACACACATTCAACCCAAATCAGTGACGGCGAAATTTGATTTATCTTTC
ATCATTGAAGAAGATCGCGATGACTATACAATCAATATCGAGTATAATACCGATTTATAT
CACTCAGAAACAGTTCGTCACATGGGTAATCAATGTATGATTATGATTGATTATATTTTG
AAGCATCAAGATACACTACAAATTTGTGATATACCAAACGGCACGGAGGAACTTCTAAAT
TGGGTCAATACGCATGTTAACGATCGAATGCTTAATGTCCCGGGAAATAAATCTATCATA
AGTTACTTTAATGAAGTTGTCTCACGACAAGGTAATCATGTTGCGCTAGTCATGAATGAT
TTGACAATGACGTATGAAACATTACGCAACTATGTGGATGCCATTGCGCACATGCTCCTA
TCAAATGGTGTGGGCAATGGTCAACGGGTTGCCTTGTTTACAGAACGTAGTTTTGAAATG
ATTGCGGCGATGTTGGCGACAGTTAAAGTAGGTGCATCTTATATACCTATCGATATTGAT
TTTCCGAATAAACGACAAGGTGCAATTTTGGAGGATGCTAAAGTAACTGCAGTCATGTCT
TACGGCGTTGAAATTGAAACGACATTACCAGTCATTCAATTGGAAAATGCTAAAGGCTTT
GTTGAATCAAAGGAAAATGAACAATATGATGATTTACATGGCAATCAACTTGAAAACACA
GCGATGTTAGATAATGAGATGTATGCTATTTACACATCTGGTACGACCGGGATGCCTAAA
GGGGTTGCCATACGACAACGAAATTTGTTGAATTTAGTGCATGCATGGTCAACTGAATTG
CAATTAGGCGACAATGAAGTATTTTTGCAACATGCAAATATTGTTTTTGATGCATCAGTT
ATGGAGATTTATTGTTGTTTGTTAAATGGTCATACGCTTGTGATTCCAGATAGAGAGGAA
CGTGTTAATCCAGAACAGTTACAACAACTCATTAATAAGCATCGTGTGACGGTTGCGTCG
ATTCCGTTACAGATGTGTAGTGTTATGGAAGACTTTTATATTGAAAAGTTGATTACAGGC
GGGGCAACTAGTACGGCATCCTTTGTTAAATATATTGAGAAGCATTGTGGCACGTATTTC
AATGCCTATGGACCATCTGAGTCAACAGTCATCACATCGTATTGGTCACATCATTGTGGT
GATTTGATACCTGAGACGATTCCAATTGGCAAACCCTTATCTAACATCCAAGTGTATATT
ATGTCAGATGGTTTGTTATGCGGTATTGGTATGCCAGGCGAGTTGTGTATTGCAGGTGAT
AGTTTAGCGATAGGATATATTAATCGTCCAGAATTAATGGCTGATAAATGGCAAAATAAT
CCATTTGGTAAAGGAAAGTTGTATCATAGTGGTGATTTAGCACGTTATACATCTGATGGT
CAAATTGAATTTTTAGGAAGAATAGATAAACAAGTGAAAGTTAACGGGTACCGTATTGAA
CTTGATGAAATTGAAAATGCAATATTAGCTATTCGTGGTATATCTGATTGTGTTGTAACA
GTAAGTCACTTTGATACGCATGATATATTGAATGCTTATTATGTCGGAGAGCAACAAGTG
GAACAGGATTTGAAGCAATATTTAAATGATCAGCTGCCTAAGTATATGATTCCTAAGACT
ATAACGCATATCGATTGTATGCCATTAACCACGAATGACAAGGTGGATACTACGCGTTTG
CCAAATCCATCACCTATACAACAGTCTAATAAAGTGTATAGCGAACCCTCTAATGAAATT
GAGCAGACATTTGTTGATGTATTTGGAGAGGTATTGAAACAAAATGATGTCGGTGTTGAC
GATGATTTCTTTGAACTTGGTGGTAACTCATTAGAGGCGATGTTAGTTGTCTCGCATTTA
AAACGATTTGGCCATCATATTTCAATGCAGACATTATACCAATATAAAACCGTGCGACAG
ATTGTTAATTATATGTACCAAAATCAACAATCATTAGTTGCATTACCGGATAATCTTTCG
GAATTACAAAAGATTGTTATGTCTCGTTATAACTTGGGTATTTTAGAGGATAGTCTAAGT
CATCGACCTCTAGGAAATACACTATTGACTGGCGCGACAGGTTTTTTAGGTGCTTATCTG
ATTGAAGTACTACAAGGATACAGTCATCGCATTTATTGTTTCATACGTGCTGATAATGAG
GAAATAGCATGGTATAAGTTGATGACGAATTTAAATGATTATTTTTCAGAAGAGACGGTT
GAAATAATGTTATCAAACATTGAAGTCATTGTTGGTGATTTCGAGTGTATGGATGATGTT
GTTTTACCAGAAAACATGGATACGATTATTCATGCAGGTGCTCGTACAGATCACTTTGGT
GATGATGATGAATTTGAAAAAGTAAATGTTCAAGGTACTGTTGATGTCATACGTTTGGCA
CAACAACATCATGCAAGGTTAATATATGTGTCTACGATAAGTGTGGGAACTTATTTTGAT
ATAGACACAGAAGATGTGACATTTTCAGAAGCGGATGTCTATAAAGGGCAACTACTAACA
TCACCATATACACGGAGCAAATTTTATAGTGAATTAAAAGTATTAGAAGCTGTAAATAAT
GGCTTAGATGGTCGGATTGTACGTGTTGGTAATTTGACGAATCCTTACAATGGAAGATGG
CATATGAGAAATATAAAGACTAACCGTTTTTCAATGGTAATGAATGATTTGTTACAACTG
GATTGTATCGGGGTTAGCATGGCTGAAATGCCTGTAGATTTTTCTTTTGTGGATACGACT
GCAAGACAAATTGTCGCATTAGCACAGGTCAACACACCACAAATCATTTACCATGTGCTA
TCACCTAATAAAATGCCGGTGAAATCTTTGTTAGAATGCGTTAAGCGCAAAGAAATTGAA
CTCGTCAGCGATGAATCATTTAATGAAATTTTACAGAAACAAGACATGTACGAAACGATT
GGATTAACTAGTGTTGACCGTGAACAACAACTAGCAATGATAGATACAACATTAACATTA
AAAATAATGAATCACATCAGTGAAAAATGGCCAACGATAACTAACAATTGGCTGTATCAT
TGGGCACAATATATCAAAACAATATTCAATAAGTAA60
120
180
240
300
360
420
480
540
600
660
720
780
840
900
960
1020
1080
1140
1200
1260
1320
1380
1440
1500
1560
1620
1680
1740
1800
1860
1920
1980
2040
2100
2160
2220
2280
2340
2400
2460
2520
2580
2640
2700
2760
2820
2880
2940
3000
3060
3120
3180
3240
3300
3360
3420
3480
3540
3600
3660
3720
3780
3840
3900
3960
4020
4080
4140
4200
4260
4320
4380
4440
4500
4560
4620
4680
4740
4800
4860
4920
4980
5040
5100
5160
5220
5280
5340
5400
5460
5520
5580
5640
5700
5760
5820
5880
5940
6000
6060
6120
6180
6240
6300
6360
6420
6480
6540
6600
6660
6720
6780
6840
6900
6960
7020
7080
7140
7176
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SAOUHSC_00144
- symbol: SAOUHSC_00144
- description: hypothetical protein
- length: 2391
- theoretical pI: 5.2802
- theoretical MW: 273481
- GRAVY: -0.23601
⊟Function[edit | edit source]
- TIGRFAM: amino acid adenylation domain (TIGR01733; HMM-score: 742.4)and 18 moreCell envelope Biosynthesis and degradation of murein sacculus and peptidoglycan D-alanine--poly(phosphoribitol) ligase, subunit 1 (TIGR01734; EC 6.1.1.13; HMM-score: 519.7)L-aminoadipate-semialdehyde dehydrogenase (TIGR03443; EC 1.2.1.31; HMM-score: 465.6)acyl-CoA ligase (AMP-forming), exosortase A-associated (TIGR03098; HMM-score: 379.1)thioester reductase domain (TIGR01746; HMM-score: 326.5)Biosynthesis of cofactors, prosthetic groups, and carriers Menaquinone and ubiquinone O-succinylbenzoate-CoA ligase (TIGR01923; EC 6.2.1.26; HMM-score: 272.5)benzoate-CoA ligase family (TIGR02262; HMM-score: 172.3)acetate--CoA ligase (TIGR02188; EC 6.2.1.1; HMM-score: 159.8)Transport and binding proteins Cations and iron carrying compounds (2,3-dihydroxybenzoyl)adenylate synthase (TIGR02275; EC 2.7.7.58; HMM-score: 135.3)cyclohexanecarboxylate-CoA ligase (TIGR03208; EC 6.2.1.-; HMM-score: 118)Central intermediary metabolism Other acetoacetate-CoA ligase (TIGR01217; EC 6.2.1.16; HMM-score: 104.4)dicarboxylate--CoA ligase PimA (TIGR03205; EC 6.2.1.23; HMM-score: 78.9)propionate--CoA ligase (TIGR02316; EC 6.2.1.17; HMM-score: 73.7)hopanoid-associated sugar epimerase (TIGR03466; HMM-score: 41)Cell envelope Biosynthesis and degradation of surface polysaccharides and lipopolysaccharides dTDP-4-dehydrorhamnose reductase (TIGR01214; EC 1.1.1.133; HMM-score: 29.1)NAD dependent epimerase/dehydratase, LLPSF_EDH_00030 family (TIGR04180; HMM-score: 25.3)4-coumarate--CoA ligase (TIGR02372; EC 6.2.1.12; HMM-score: 24.5)Hypothetical proteins Conserved TIGR01777 family protein (TIGR01777; HMM-score: 15.4)Energy metabolism Electron transport archaeoflavoprotein AfpA (TIGR02699; HMM-score: 13.3)
- TheSEED :
- Long-chain-fatty-acid--CoA ligase (EC 6.2.1.3)
- PFAM: ANL (CL0378) AMP-binding; AMP-binding enzyme (PF00501; HMM-score: 492)and 9 moreCoA-acyltrans (CL0149) Condensation; Condensation domain (PF00668; HMM-score: 338.4)NADP_Rossmann (CL0063) NAD_binding_4; Male sterility protein (PF07993; HMM-score: 107.5)PP-binding (CL0314) PP-binding; Phosphopantetheine attachment site (PF00550; HMM-score: 98.3)AMP-binding_C (CL0531) AMP-binding_C; AMP-binding enzyme C-terminal domain (PF13193; HMM-score: 61.3)NADP_Rossmann (CL0063) Epimerase; NAD dependent epimerase/dehydratase family (PF01370; HMM-score: 61.1)3Beta_HSD; 3-beta hydroxysteroid dehydrogenase/isomerase family (PF01073; HMM-score: 45.5)GDP_Man_Dehyd; GDP-mannose 4,6 dehydratase (PF16363; HMM-score: 40)RmlD_sub_bind; RmlD substrate binding domain (PF04321; HMM-score: 32.5)Polysacc_synt_2; Polysaccharide biosynthesis protein (PF02719; HMM-score: 21.6)
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors:
- effectors:
⊟Localization[edit | edit source]
- PSORTb: Cytoplasmic
- Cytoplasmic Score: 9.97
- Cytoplasmic Membrane Score: 0
- Cellwall Score: 0.01
- Extracellular Score: 0.02
- Internal Helices: 0
- DeepLocPro: Cytoplasmic
- Cytoplasmic Score: 0.9339
- Cytoplasmic Membrane Score: 0.054
- Cell wall & surface Score: 0.0011
- Extracellular Score: 0.011
- LocateP: Intracellular
- Prediction by SwissProt Classification: Cytoplasmic
- Pathway Prediction: No pathway
- Intracellular possibility: 1
- Signal peptide possibility: -1
- N-terminally Anchored Score: 1
- Predicted Cleavage Site: No CleavageSite
- SignalP: no predicted signal peptide
- SP(Sec/SPI): 0.003937
- TAT(Tat/SPI): 0.000274
- LIPO(Sec/SPII): 0.000633
- predicted transmembrane helices (TMHMM): 0
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MIMGNLRFQQEYFRIYKNNTESTTHRNAYWVKLAKNVEATKMMYALSTIVQQHASIRHFFDVTTDDNLTMILHEFLPFIEIKQVPSSSANYDLEAFFKQELSTYHFNDSPLFKVKLFQFADAAYILLDFHVSIFDDSQIDIFLDDLCNAYRGNTVINNTRQHAHINRNDDKDNQDASHIALDSNYFRLENNSDIHIDSYFPIKHPFEQALYQTYLIDDMTSIDMASLAVSVYLANHIMSQQHDVTLGIHVPSHLPNDLHGNIVPLTLTIDAKDVCQRFTTDFNKCVLQNMSQLQCAKSSLSLETIFHCYHHMMSCCNDVIEDVHQIHDAHTSLADIEIFPHQHGFKIIYNSAAYDLLSIETLSDLVRNIYLQITEENGNKRTTVDELNLMTERDIQLYDDINLSLPEIDDAQTVVTLFEQQVEATPNHVAVQFDGVFITYQTLNARANDLAHRLRNQYGVEPNDRVAVIAEKSIEMIIAMIGVLKAGGAYVPIDPNYPSDRQEYILKDVTPKVVITYQALYENGKQNINHIDLNKIAWKNIDNLSKCNTLEDHAYVIYTSGTTGNPKGTLIPHRGIVRLVHQNHYVPLNEETTILLSGTIAFDAATFEIYGALLNGGKLIVAKKEQLLNPIAVEQLINENDVNTMWLTSSLFNQIASERIEVLVPLKYLLIGGEVLNAKWVDLLNQKPKHPQIINGYGPTENTTFTTTYNIPNKVPNRIPIGKPILGTHVYIMQGERRCGVGIPGELCTSGFGLAAGYLNQPELTADKFIKDSNINQLMYRSGDIVRLLPDGNIDYLYRKDKQVKIRGFRIELSEVEHALERIQGINKAVVIVQNHDQDQYIVAYYEAMHTLSHNKIKSQLRMTLPEYMIPVNFMHIEQIPITINGKLDKKALPIMDYVDTDAYVAPSTDTEHLLCQIFADILHVNQVGIHDNFFELGGHSLKATLVVNRIEASTGKRLQIGDLLQKPTVFELAQAIAKVQEQNYEVIPETIVKDDYVLSSAQKRMYLLWKSNHKDTVYNVPFLWRLSSELNVAQLRQAVQRLIARHEILRTQYIVVDDEVRQRIVADVAVDFEEVNTHFTDEQEIMRQFVAPFNLEKPSQIRVRYIRSPLHAYLFIDTHHIINDGMSNIQLMNDLNALYQHKLLLPLKLQYKDYSEWMSHRDMTKHRQYWLSQFKDEVPILSLPTDYVRPNIKTTNGAMMSFTMNQQMRQLLQKYVEKHQITDFMFFMSVVMTLLSRYARKDDVVVGSVMSARMHKGTEQMLGMFANTLVYRGQPSPDKMWTQFLQEVKEMSLEAYEHQEYPFECLVNDLDQSHDASRNPLFDVMLVLQNNETNHAHFGHSKLTHIQPKSVTAKFDLSFIIEEDRDDYTINIEYNTDLYHSETVRHMGNQCMIMIDYILKHQDTLQICDIPNGTEELLNWVNTHVNDRMLNVPGNKSIISYFNEVVSRQGNHVALVMNDLTMTYETLRNYVDAIAHMLLSNGVGNGQRVALFTERSFEMIAAMLATVKVGASYIPIDIDFPNKRQGAILEDAKVTAVMSYGVEIETTLPVIQLENAKGFVESKENEQYDDLHGNQLENTAMLDNEMYAIYTSGTTGMPKGVAIRQRNLLNLVHAWSTELQLGDNEVFLQHANIVFDASVMEIYCCLLNGHTLVIPDREERVNPEQLQQLINKHRVTVASIPLQMCSVMEDFYIEKLITGGATSTASFVKYIEKHCGTYFNAYGPSESTVITSYWSHHCGDLIPETIPIGKPLSNIQVYIMSDGLLCGIGMPGELCIAGDSLAIGYINRPELMADKWQNNPFGKGKLYHSGDLARYTSDGQIEFLGRIDKQVKVNGYRIELDEIENAILAIRGISDCVVTVSHFDTHDILNAYYVGEQQVEQDLKQYLNDQLPKYMIPKTITHIDCMPLTTNDKVDTTRLPNPSPIQQSNKVYSEPSNEIEQTFVDVFGEVLKQNDVGVDDDFFELGGNSLEAMLVVSHLKRFGHHISMQTLYQYKTVRQIVNYMYQNQQSLVALPDNLSELQKIVMSRYNLGILEDSLSHRPLGNTLLTGATGFLGAYLIEVLQGYSHRIYCFIRADNEEIAWYKLMTNLNDYFSEETVEIMLSNIEVIVGDFECMDDVVLPENMDTIIHAGARTDHFGDDDEFEKVNVQGTVDVIRLAQQHHARLIYVSTISVGTYFDIDTEDVTFSEADVYKGQLLTSPYTRSKFYSELKVLEAVNNGLDGRIVRVGNLTNPYNGRWHMRNIKTNRFSMVMNDLLQLDCIGVSMAEMPVDFSFVDTTARQIVALAQVNTPQIIYHVLSPNKMPVKSLLECVKRKEIELVSDESFNEILQKQDMYETIGLTSVDREQQLAMIDTTLTLKIMNHISEKWPTITNNWLYHWAQYIKTIFNK
⊟Experimental data[edit | edit source]
- experimentally validated: PeptideAtlas [1] [2]
- protein localization: data available for COL
- quantitative data / protein copy number per cell:
- interaction partners:
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
⊟Regulation[edit | edit source]
- data available for JSNZ
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: [3]
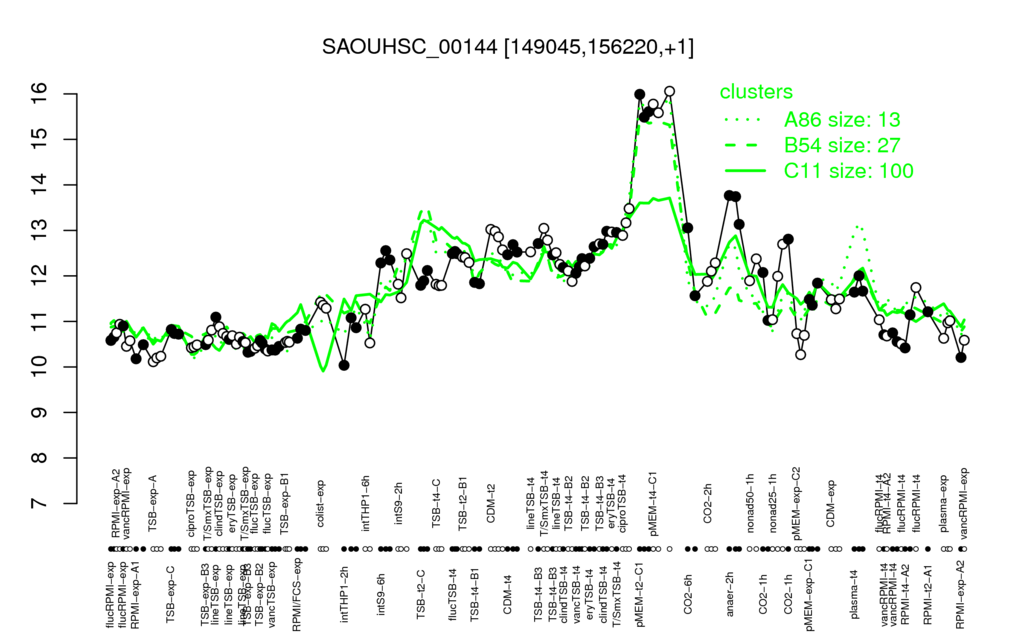 Multi-gene expression profiles
Multi-gene expression profiles
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
- Aureolib: no data available
⊟Protein stability[edit | edit source]
- half-life: no data available
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You can add further information about the gene and protein here. [edit]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ Maren Depke, Stephan Michalik, Alexander Rabe, Kristin Surmann, Lars Brinkmann, Nico Jehmlich, Jörg Bernhardt, Michael Hecker, Bernd Wollscheid, Zhi Sun, Robert L Moritz, Uwe Völker, Frank Schmidt
A peptide resource for the analysis of Staphylococcus aureus in host-pathogen interaction studies.
Proteomics: 2015, 15(21);3648-61
[PubMed:26224020] [WorldCat.org] [DOI] (I p) - ↑ Stephan Michalik, Maren Depke, Annette Murr, Manuela Gesell Salazar, Ulrike Kusebauch, Zhi Sun, Tanja C Meyer, Kristin Surmann, Henrike Pförtner, Petra Hildebrandt, Stefan Weiss, Laura Marcela Palma Medina, Melanie Gutjahr, Elke Hammer, Dörte Becher, Thomas Pribyl, Sven Hammerschmidt, Eric W Deutsch, Samuel L Bader, Michael Hecker, Robert L Moritz, Ulrike Mäder, Uwe Völker, Frank Schmidt
A global Staphylococcus aureus proteome resource applied to the in vivo characterization of host-pathogen interactions.
Sci Rep: 2017, 7(1);9718
[PubMed:28887440] [WorldCat.org] [DOI] (I e) - ↑ 3.0 3.1 3.2 Ulrike Mäder, Pierre Nicolas, Maren Depke, Jan Pané-Farré, Michel Debarbouille, Magdalena M van der Kooi-Pol, Cyprien Guérin, Sandra Dérozier, Aurelia Hiron, Hanne Jarmer, Aurélie Leduc, Stephan Michalik, Ewoud Reilman, Marc Schaffer, Frank Schmidt, Philippe Bessières, Philippe Noirot, Michael Hecker, Tarek Msadek, Uwe Völker, Jan Maarten van Dijl
Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions.
PLoS Genet: 2016, 12(4);e1005962
[PubMed:27035918] [WorldCat.org] [DOI] (I e)
