Jump to navigation
Jump to search
NCBI: 03-AUG-2016
⊟Summary[edit | edit source]
- organism: Staphylococcus aureus NCTC8325
- locus tag: SAOUHSC_00545
- pan locus tag?: SAUPAN002336000
- symbol: SAOUHSC_00545
- pan gene symbol?: sdrD
- synonym:
- product: fibrinogen-binding protein SdrD
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
- type: CDS
- locus tag: SAOUHSC_00545
- symbol: SAOUHSC_00545
- product: fibrinogen-binding protein SdrD
- replicon: chromosome
- strand: +
- coordinates: 551105..555154
- length: 4050
- essential: no DEG other strains
⊟Accession numbers[edit | edit source]
- Gene ID: 3920825 NCBI
- RefSeq: YP_499118 NCBI
- BioCyc: G1I0R-516 BioCyc
- MicrobesOnline: 1289028 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421
481
541
601
661
721
781
841
901
961
1021
1081
1141
1201
1261
1321
1381
1441
1501
1561
1621
1681
1741
1801
1861
1921
1981
2041
2101
2161
2221
2281
2341
2401
2461
2521
2581
2641
2701
2761
2821
2881
2941
3001
3061
3121
3181
3241
3301
3361
3421
3481
3541
3601
3661
3721
3781
3841
3901
3961
4021ATGCTAAACAGAGAAAATAAAACGGCAATAACAAGGAAAGGCATGGTATCCAATCGATTA
AATAAATTTTCGATTAGAAAGTACACAGTGGGAACAGCATCAATTTTAGTAGGTACAACA
TTAATTTTTGGTCTGGGGAACCAAGAAGCAAAGGCTGCAGAAAGTACTAATAAAGAATTG
AACGAAGCGACAACTTCAGCAAGTGATAATCAATCGAGTGATAAAGTTGATATGCAGCAA
CTAAATCAAGAAGACAATACTAAAAATGATAATCAAAAAGAAATGGTATCATCTCAAGGT
AATGAAACGACTTCAAATGGGAATAAATTAATAGAAAAAGAAAGTGTACAATCTACCACT
GGAAATAAAGTTGAAGTTTCAACTGCCAAATCAGATGAGCAAGCTTCACCAAAATCTACG
AATGAAGATTTAAACACTAAACAAACTATAAGTAATCAAGAAGCGTTACAACCTGATTTG
CAAGAGAATAAATCAGTGGTAAATGTTCAACCAACTAATGAGGAAAACAAAAAGGTAGAT
GCCAAAACTGAATCAACTACATTAAATGTTAAAAGTGATGCTATCAAGAGTAATGATGAA
ACTCTTGTTGATAACAATAGTAATTCAAATAATGAAAATAATGCAGATATCATTTTGCCA
AAAAGTACAGCACCTAAACGTTTGAATACAAGAATGCGTATAGCAGCAGTACAGCCATCA
TCAACAGAGGCTAAAAATGTTAATGATTTAATCACATCAAATACAACATTAACTGTCGTT
GATGCAGATAAAAACAATAAAATCGTACCAGCCCAAGATTATTTATCATTAAAATCACAA
ATTACAGTTGATGACAAAGTTAAATCAGGTGATTATTTCACAATTAAATACTCAGATACA
GTACAAGTATATGGATTGAATCCGGAAGATATTAAAAATATTGGTGATATTAAAGATCCA
AATAATGGTGAAACAATTGCGACTGCAAAACATGATACTGCAAATAATTTAATTACATAT
ACATTTACAGATTATGTTGATCGATTTAATTCTGTACAAATGGGAATTAATTATTCAATT
TATATGGATGCTGATACAATTCCTGTTAGTAAAAACGATGTTGAGTTTAATGTTACGATA
GGTAATACTACAACAAAAACAACTGCTAACATTCAATATCCAGATTATGTTGTAAATGAG
AAAAATTCAATTGGATCAGCGTTCACTGAAACAGTTTCACATGTTGGAAATAAAGAAAAT
CCAGGGTACTATAAACAAACGATTTATGTAAATCCATCGGAAAATTCTTTAACAAATGCC
AAACTAAAAGTTCAAGCTTACCACTCAAGTTATCCTAATAATATCGGGCAAATAAATAAA
GATGTAACAGATATAAAAATATATCAAGTTCCTAAAGGTTATACATTAAATAAAGGATAC
GATGTGAATACTAAAGAGCTTACAGATGTAACAAATCAATACTTGCAGAAAATTACATAT
GGCGACAACAATAGCGCTGTTATTGATTTTGGAAATGCAGATTCTGCTTATGTTGTAATG
GTTAATACAAAATTCCAATATACAAATAGCGAAAGCCCAACACTTGTTCAAATGGCTACT
TTATCTTCAACAGGTAATAAATCCGTTTCTACTGGCAATGCTTTAGGATTTACTAATAAC
CAAAGTGGCGGAGCTGGTCAAGAAGTATATAAAATTGGTAACTACGTATGGGAAGATACT
AATAAAAACGGTGTTCAAGAATTAGGAGAAAAAGGCGTTGGCAATGTAACTGTAACTGTA
TTTGATAATAATACAAATACAAAAGTAGGAGAAGCAGTTACTAAAGAAGATGGGTCATAC
TTGATTCCAAACTTACCTAATGGAGATTACCGTGTAGAATTTTCAAACTTACCAAAAGGT
TATGAAGTAACCCCTTCAAAACAAGGTAATAACGAAGAATTAGATTCAAACGGCTTATCT
TCAGTTATTACAGTTAATGGCAAAGATAACTTATCTGCAGACTTAGGTATTTACAAACCT
AAATACAACTTAGGTGACTATGTCTGGGAAGATACAAATAAAAATGGTATCCAAGACCAA
GATGAAAAAGGTATATCTGGCGTAACGGTAACATTAAAAGATGAAAACGGTAACGTGTTA
AAAACAGTTACAACAGACGCTGATGGCAAATATAAATTTACTGATTTAGATAATGGTAAT
TATAAAGTTGAATTTACTACACCAGAAGGCTATACACCGACTACAGTAACATCTGGTAGC
GACATTGAAAAAGACTCTAATGGTTTAACAACAACAGGTGTTATTAATGGTGCTGATAAC
ATGACATTAGATAGTGGATTCTACAAAACACCAAAATATAATTTAGGTAATTATGTATGG
GAAGATACAAATAAAGATGGTAAGCAGGATTCAACTGAAAAAGGTATTTCAGGCGTAACA
GTTACATTGAAAAATGAAAACGGTGAAGTTTTACAAACAACTAAAACAGATAAAGATGGT
AAATATCAATTTACTGGATTAGAAAATGGAACTTATAAAGTTGAATTCGAAACACCATCA
GGTTACACACCAACACAAGTAGGTTCAGGAACTGATGAAGGTATAGATTCAAATGGTACA
TCAACAACAGGTGTCATTAAAGATAAAGATAACGATACTATTGACTCTGGTTTCTACAAA
CCGACTTACAACTTAGGTGACTATGTATGGGAAGATACAAATAAAAACGGTGTTCAAGAT
AAAGATGAAAAGGGCATTTCAGGTGTAACAGTTACGTTAAAAGATGAAAACGACAAAGTT
TTAAAAACAGTTACAACAGATGAAAATGGTAAATATCAATTCACTGATTTAAACAATGGA
ACTTATAAAGTTGAATTCGAGACACCATCAGGTTATACACCAACTTCAGTAACTTCTGGA
AATGATACTGAAAAAGATTCTAATGGTTTAACAACAACAGGTGTCATTAAAGATGCAGAT
AACATGACATTAGACAGTGGTTTCTATAAAACACCAAAATATAGTTTAGGTGATTATGTT
TGGTACGACAGTAATAAAGACGGCAAACAAGATTCAACTGAAAAAGGTATCAAAGATGTT
AAAGTTACTTTATTAAATGAAAAAGGCGAAGTAATTGGAACAACTAAAACAGATGAAAAT
GGTAAATACTGCTTTGATAATTTAGATAGCGGTAAATACAAAGTTATTTTTGAAAAGCCT
GCTGGCTTAACACAAACAGTTACAAATACAACTGAAGATGATAAAGATGCAGATGGTGGC
GAAGTTGACGTAACAATTACGGATCATGATGATTTCACACTTGATAACGGATACTTCGAA
GAAGATACATCAGACAGCGATTCAGACTCAGATAGTGACTCAGACAGCGACTCAGACTCA
GACAGCGACTCAGACTCAGACAGTGATTCAGATTCAGACAGCGACTCAGATTCAGATAGC
GACTCAGATTCGGACAGCGATTCAGACTCAGATAGCGACTCAGATTCAGATAGCGATTCA
GACTCAGACAGCGACTCAGATTCAGATAGCGATTCGGACTCAGACAGCGATTCAGACTCA
GATAGCGACTCAGACTCAGACAGCGACTCAGATTCAGATAGCGATTCGGACTCAGATAGC
GACTCAGATTCAGACAGCGATTCAGACTCAGATAGCGACTCAGATTCAGACAGCGATTCA
GACTCAGATAGCGACTCAGACTCAGACAGTGATTCAGATTCAGACAGCGACTCAGACTCA
GATAGCGACTCAGATTCGGACAGCGACTCAGACTCTGATAGCGACTCAGACTCAGACAGT
GATTCAGACAGCGATTCAGACTCGGATGCAGGAAAACATACACCTGTTAAACCAATGAGT
ACTACTAAAGACCATCACAATAAAGCAAAAGCATTACCAGAAACAGGTAGTGAAAATAAC
GGCTCAAATAACGCAACGTTATTTGGTGGATTATTTGCAGCATTAGGTTCATTATTGTTA
TTCGGTCGTCGCAAAAAACAAAACAAATAA60
120
180
240
300
360
420
480
540
600
660
720
780
840
900
960
1020
1080
1140
1200
1260
1320
1380
1440
1500
1560
1620
1680
1740
1800
1860
1920
1980
2040
2100
2160
2220
2280
2340
2400
2460
2520
2580
2640
2700
2760
2820
2880
2940
3000
3060
3120
3180
3240
3300
3360
3420
3480
3540
3600
3660
3720
3780
3840
3900
3960
4020
4050
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SAOUHSC_00545
- symbol: SAOUHSC_00545
- description: fibrinogen-binding protein SdrD
- length: 1349
- theoretical pI: 3.9647
- theoretical MW: 146086
- GRAVY: -1.00586
⊟Function[edit | edit source]
- TIGRFAM: gram-positive signal peptide, YSIRK family (TIGR01168; HMM-score: 49)and 2 moreEnergy metabolism Other protocatechuate 3,4-dioxygenase, beta subunit (TIGR02422; EC 1.13.11.3; HMM-score: 17.5)Cell envelope Other LPXTG cell wall anchor domain (TIGR01167; HMM-score: 16.6)
- TheSEED :
- Adhesin of unknown specificity SdrD
- PFAM: Transthyretin (CL0287) SdrD_B; SdrD B-like domain (PF17210; HMM-score: 496.9)and 30 moreCarboxypepD_reg; Carboxypeptidase regulatory-like domain (PF13620; HMM-score: 177.8)Adhesin (CL0204) SdrG_C_C; C-terminus of bacterial fibrinogen-binding adhesin (PF10425; HMM-score: 133.1)Transthyretin (CL0287) SpaA; Prealbumin-like fold domain (PF17802; HMM-score: 116.6)E-set (CL0159) Big_8; Bacterial Ig domain (PF17961; HMM-score: 88.4)bMG3; Bacterial alpha-2-macroglobulin MG3 domain (PF11974; HMM-score: 84.9)Transthyretin (CL0287) CarbopepD_reg_2; CarboxypepD_reg-like domain (PF13715; HMM-score: 69.3)EMC7_beta-sandw; ER membrane protein complex subunit 7, beta-sandwich domain (PF09430; HMM-score: 61.6)Ig_DR_A0283-like; DR_A0283-like, Ig-like domain (PF24025; HMM-score: 59.6)MetallophosN; N terminal of Calcineurin-like phosphoesterase (PF16371; HMM-score: 54.5)NOMO1-like_9th; BOS complex subunit NOMO1-like, beta sandwich domain (PF22902; HMM-score: 50.8)no clan defined YSIRK_signal; YSIRK type signal peptide (PF04650; HMM-score: 46.1)Transthyretin (CL0287) NOMO1-like_1st; BOS complex subunit NOMO1-like, first beta sandwich domain (PF22898; HMM-score: 40.1)no clan defined Gram_pos_anchor; LPXTG cell wall anchor motif (PF00746; HMM-score: 38.4)E-set (CL0159) Ig_J; Tip attachment protein J, Ig-like domain (PF24421; HMM-score: 35)Transthyretin (CL0287) DUF1416; Protein of unknown function (DUF1416) (PF07210; HMM-score: 32.8)E-set (CL0159) Big_6; Bacterial Ig domain (PF17936; HMM-score: 31.1)Transthyretin (CL0287) TTR-52; Transthyretin-like family (PF01060; HMM-score: 29.8)GOLD-like (CL0521) EMP24_GP25L; emp24/gp25L/p24 family/GOLD (PF01105; HMM-score: 27.6)Transthyretin (CL0287) NOMO_3rd; NOMO third transthyretin-like domain (PF23193; HMM-score: 26.8)NOMO_5th; NOMO fifth transthyretin-like domain (PF23194; HMM-score: 26.5)NOMO1-like_2nd; BOS complex subunit NOMO1-like, second beta sandwich domain (PF22904; HMM-score: 26.1)Dioxygenase_C; Dioxygenase (PF00775; HMM-score: 23.7)E-set (CL0159) MG4; Macroglobulin domain MG4 (PF17789; HMM-score: 20.2)SH3 (CL0010) DUF4926; Domain of unknown function (DUF4926) (PF16277; HMM-score: 16.8)Transthyretin (CL0287) fn3_3; Polysaccharide lyase family 4, domain II (PF14686; HMM-score: 15.4)DUF2606; Protein of unknown function (DUF2606) (PF10794; HMM-score: 14.6)no clan defined ArgL-like; ArgL-like (PF22864; HMM-score: 14.5)E-set (CL0159) Y_Y_Y; Y_Y_Y domain (PF07495; HMM-score: 14)bBprotInhib (CL0354) Inh; Protease inhibitor Inh (PF02974; HMM-score: 12.9)HSP20 (CL0190) HSP20; Hsp20/alpha crystallin family (PF00011; HMM-score: 12)
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors:
- effectors:
⊟Localization[edit | edit source]
- PSORTb: Cellwall
- Cytoplasmic Score: 0
- Cytoplasmic Membrane Score: 0
- Cellwall Score: 10
- Extracellular Score: 0
- Internal Helix: 1
- DeepLocPro: Cell wall & surface
- Cytoplasmic Score: 0
- Cytoplasmic Membrane Score: 0.0007
- Cell wall & surface Score: 0.8827
- Extracellular Score: 0.1166
- LocateP: LPxTG Cell-wall anchored
- Prediction by SwissProt Classification: Cell Wall
- Pathway Prediction: Sec-(SPI)
- Intracellular possibility: 0
- Signal peptide possibility: 1
- N-terminally Anchored Score: -2
- Predicted Cleavage Site: found LPxTG motif :LPETG
- SignalP: Signal peptide SP(Sec/SPI) length 52 aa
- SP(Sec/SPI): 0.812615
- TAT(Tat/SPI): 0.093189
- LIPO(Sec/SPII): 0.002787
- Cleavage Site: CS pos: 52-53. AKA-AE. Pr: 0.8678
- predicted transmembrane helices (TMHMM): 1
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MLNRENKTAITRKGMVSNRLNKFSIRKYTVGTASILVGTTLIFGLGNQEAKAAESTNKELNEATTSASDNQSSDKVDMQQLNQEDNTKNDNQKEMVSSQGNETTSNGNKLIEKESVQSTTGNKVEVSTAKSDEQASPKSTNEDLNTKQTISNQEALQPDLQENKSVVNVQPTNEENKKVDAKTESTTLNVKSDAIKSNDETLVDNNSNSNNENNADIILPKSTAPKRLNTRMRIAAVQPSSTEAKNVNDLITSNTTLTVVDADKNNKIVPAQDYLSLKSQITVDDKVKSGDYFTIKYSDTVQVYGLNPEDIKNIGDIKDPNNGETIATAKHDTANNLITYTFTDYVDRFNSVQMGINYSIYMDADTIPVSKNDVEFNVTIGNTTTKTTANIQYPDYVVNEKNSIGSAFTETVSHVGNKENPGYYKQTIYVNPSENSLTNAKLKVQAYHSSYPNNIGQINKDVTDIKIYQVPKGYTLNKGYDVNTKELTDVTNQYLQKITYGDNNSAVIDFGNADSAYVVMVNTKFQYTNSESPTLVQMATLSSTGNKSVSTGNALGFTNNQSGGAGQEVYKIGNYVWEDTNKNGVQELGEKGVGNVTVTVFDNNTNTKVGEAVTKEDGSYLIPNLPNGDYRVEFSNLPKGYEVTPSKQGNNEELDSNGLSSVITVNGKDNLSADLGIYKPKYNLGDYVWEDTNKNGIQDQDEKGISGVTVTLKDENGNVLKTVTTDADGKYKFTDLDNGNYKVEFTTPEGYTPTTVTSGSDIEKDSNGLTTTGVINGADNMTLDSGFYKTPKYNLGNYVWEDTNKDGKQDSTEKGISGVTVTLKNENGEVLQTTKTDKDGKYQFTGLENGTYKVEFETPSGYTPTQVGSGTDEGIDSNGTSTTGVIKDKDNDTIDSGFYKPTYNLGDYVWEDTNKNGVQDKDEKGISGVTVTLKDENDKVLKTVTTDENGKYQFTDLNNGTYKVEFETPSGYTPTSVTSGNDTEKDSNGLTTTGVIKDADNMTLDSGFYKTPKYSLGDYVWYDSNKDGKQDSTEKGIKDVKVTLLNEKGEVIGTTKTDENGKYCFDNLDSGKYKVIFEKPAGLTQTVTNTTEDDKDADGGEVDVTITDHDDFTLDNGYFEEDTSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDAGKHTPVKPMSTTKDHHNKAKALPETGSENNGSNNATLFGGLFAALGSLLLFGRRKKQNK
⊟Experimental data[edit | edit source]
- experimentally validated: PeptideAtlas [1] [2]
- protein localization: data available for COL
- quantitative data / protein copy number per cell:
- interaction partners:
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
- predicted SigA promoter [3] : S197 > SAOUHSC_00545
⊟Regulation[edit | edit source]
- regulator:
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: [3]
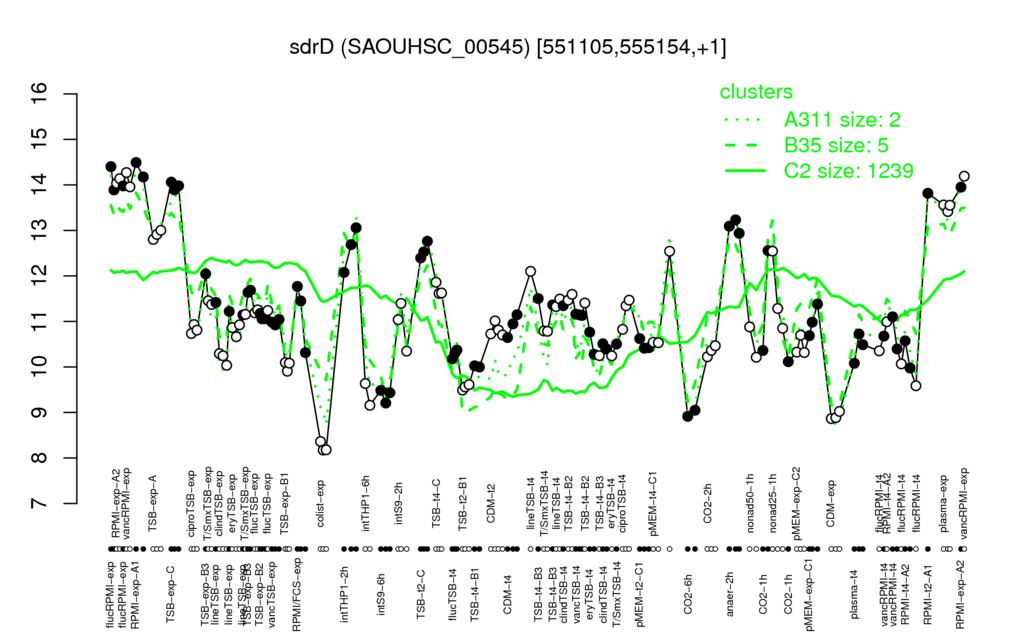 Multi-gene expression profiles
Multi-gene expression profiles
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
- Aureolib: no data available
⊟Protein stability[edit | edit source]
- half-life: no data available
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You can add further information about the gene and protein here. [edit]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ Maren Depke, Stephan Michalik, Alexander Rabe, Kristin Surmann, Lars Brinkmann, Nico Jehmlich, Jörg Bernhardt, Michael Hecker, Bernd Wollscheid, Zhi Sun, Robert L Moritz, Uwe Völker, Frank Schmidt
A peptide resource for the analysis of Staphylococcus aureus in host-pathogen interaction studies.
Proteomics: 2015, 15(21);3648-61
[PubMed:26224020] [WorldCat.org] [DOI] (I p) - ↑ Stephan Michalik, Maren Depke, Annette Murr, Manuela Gesell Salazar, Ulrike Kusebauch, Zhi Sun, Tanja C Meyer, Kristin Surmann, Henrike Pförtner, Petra Hildebrandt, Stefan Weiss, Laura Marcela Palma Medina, Melanie Gutjahr, Elke Hammer, Dörte Becher, Thomas Pribyl, Sven Hammerschmidt, Eric W Deutsch, Samuel L Bader, Michael Hecker, Robert L Moritz, Ulrike Mäder, Uwe Völker, Frank Schmidt
A global Staphylococcus aureus proteome resource applied to the in vivo characterization of host-pathogen interactions.
Sci Rep: 2017, 7(1);9718
[PubMed:28887440] [WorldCat.org] [DOI] (I e) - ↑ 3.0 3.1 Ulrike Mäder, Pierre Nicolas, Maren Depke, Jan Pané-Farré, Michel Debarbouille, Magdalena M van der Kooi-Pol, Cyprien Guérin, Sandra Dérozier, Aurelia Hiron, Hanne Jarmer, Aurélie Leduc, Stephan Michalik, Ewoud Reilman, Marc Schaffer, Frank Schmidt, Philippe Bessières, Philippe Noirot, Michael Hecker, Tarek Msadek, Uwe Völker, Jan Maarten van Dijl
Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions.
PLoS Genet: 2016, 12(4);e1005962
[PubMed:27035918] [WorldCat.org] [DOI] (I e)
⊟Relevant publications[edit | edit source]
T J Foster, M Höök
Surface protein adhesins of Staphylococcus aureus.
Trends Microbiol: 1998, 6(12);484-8
[PubMed:10036727] [WorldCat.org] [DOI] (P p)Mark J J B Sibbald, Theresa Winter, Magdalena M van der Kooi-Pol, G Buist, E Tsompanidou, Tjibbe Bosma, Tina Schäfer, Knut Ohlsen, Michael Hecker, Haike Antelmann, Susanne Engelmann, Jan Maarten van Dijl
Synthetic effects of secG and secY2 mutations on exoproteome biogenesis in Staphylococcus aureus.
J Bacteriol: 2010, 192(14);3788-800
[PubMed:20472795] [WorldCat.org] [DOI] (I p)
