⊟Summary[edit | edit source]
- organism: Staphylococcus aureus NCTC8325
- locus tag: SAOUHSC_01187
- pan locus tag?: SAUPAN003503000
- symbol: SAOUHSC_01187
- pan gene symbol?: pknB
- synonym:
- product: hypothetical protein
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
⊟Accession numbers[edit | edit source]
- Gene ID: 3919320 NCBI
- RefSeq: YP_499726 NCBI
- BioCyc: G1I0R-1113 BioCyc
- MicrobesOnline: 1289640 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421
481
541
601
661
721
781
841
901
961
1021
1081
1141
1201
1261
1321
1381
1441
1501
1561
1621
1681
1741
1801
1861
1921
1981ATGATAGGTAAAATAATAAATGAACGATATAAAATTGTAGATAAGCTTGGCGGCGGTGGC
ATGAGTACCGTTTATCTTGCTGAAGATACGATACTTAACATTAAAGTTGCAATTAAGGCG
ATTTTTATACCACCTAGAGAAAAAGAAGAAACATTAAAACGTTTTGAACGAGAAGTACAT
AACTCATCACAGCTATCACATCAAAATATAGTAAGTATGATCGATGTTGATGAAGAAGAT
GACTGTTACTACTTAGTAATGGAATATATCGAAGGTCCGACTTTGTCTGAGTATATTGAA
AGTCATGGGCCATTAAGTGTTGACACAGCGATTAATTTTACGAATCAAATATTGGATGGC
ATTAAACATGCGCATGATATGCGTATTGTACATAGAGATATTAAGCCACAAAATATATTA
ATTGACAGCAATAAAACGTTGAAAATATTTGATTTTGGAATTGCTAAAGCTTTAAGTGAG
ACGTCTTTAACTCAGACTAATCATGTGTTAGGTACTGTGCAGTACTTTTCGCCAGAACAA
GCAAAAGGTGAGGCAACGGATGAATGTACAGATATTTATTCTATAGGTATTGTGTTATAT
GAAATGCTTGTTGGTGAACCACCCTTTAATGGAGAAACTGCAGTTAGCATTGCGATTAAA
CATATTCAGGATTCTGTGCCAAATGTGACAACAGATGTACGTAAGGATATTCCGCAATCT
TTAAGTAATGTCATTTTACGCGCTACAGAAAAAGACAAAGCGAATCGTTACAAAACAATT
CAAGAAATGAAAGATGATTTGAGTAGTGTTTTACATGAAAATCGAGCGAATGAAGATGTC
TATGAACTCGATAAAATGAAAACGATAGCGGTACCTTTGAAAAAAGAAGATCTAGCAAAG
CATATTAGTGAACATAAGTCGAATCAACCTAAACGTGAAACGACGCAAGTACCTATTGTA
AATGGGCCTGCTCATCATCAGCAATTCCAAAAGCCAGAAGGTACGGTGTACGAACCAAAA
CCTAAAAAGAAATCAACACGAAAGATTGTGCTCTTATCACTAATCTTTTCGTTGTTAATG
ATTGCACTTGTTTCTTTTGTGGCAATGGCAATGTTTGGTAATAAATACGAAGAGACACCT
GATGTAATCGGGAAATCTGTAAAAGAAGCAGAGCAAATATTCAATAAAAACAACCTGAAA
TTGGGTAAAATTTCTAGAAGTTATAGTGATAAATATCCTGAAAATGAAATTATTAAGACA
ACTCCTAATACTGGTGAACGTGTTGAACGTGGTGACAGTGTTGATGTTGTTATATCAAAG
GGCCCTGAAAAGGTTAAAATGCCAAATGTCATTGGTTTACCTAAGGAGGAAGCCTTGCAG
AAATTAAAATCGTTAGGTCTTAAAGATGTTACGATTGAAAAAGTATATAATAATCAAGCG
CCAAAAGGATACATTGCAAATCAAAGTGTAACCGCAAATACTGAAATCGCTATTCATGAT
TCTAATATTAAACTATATGAATCTTTAGGCATTAAGCAAGTTTATGTAGAAGACTTTGAA
CATAAATCCTTTAGCAAAGCTAAAAAAGCCTTAGAAGAAAAAGGGTTTAAAGTTGAAAGT
AAGGAAGAGTATAGTGACGATATTGATGAGGGTGATGTGATTTCTCAATCTCCTAAAGGA
AAATCAGTAGATGAGGGGTCAACGATTTCATTTGTTGTTTCTAAAGGTAAAAAAAGTGAC
TCATCAGATGTCAAAACGACAACTGAATCGGTAGATGTACCATACACTGGTAAAAATGAT
AAGTCACAAAAAGTTAAAGTTTATATTAAAGATAAAGATAATGACGGTTCAACTGAAAAA
GGTAGTTTCGATATTACTAGTGATCAACGTATAGACATTCCTTTAAGAATTGAAAAAGGA
AAAACAGCAAGTTATATTGTTAAAGTTGACGGTAAAACTGTAGCTGAAAAAGAAGTCAGC
TATGATGATGTATAA60
120
180
240
300
360
420
480
540
600
660
720
780
840
900
960
1020
1080
1140
1200
1260
1320
1380
1440
1500
1560
1620
1680
1740
1800
1860
1920
1980
1995
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SAOUHSC_01187
- symbol: SAOUHSC_01187
- description: hypothetical protein
- length: 664
- theoretical pI: 5.9417
- theoretical MW: 74362.8
- GRAVY: -0.546837
⊟Function[edit | edit source]
- TIGRFAM: Cellular processes Toxin production and resistance TOMM system kinase/cyclase fusion protein (TIGR03903; HMM-score: 150.6)and 3 moreUnknown function General Kae1-associated kinase Bud32 (TIGR03724; EC 2.7.-.-; HMM-score: 41.9)Biosynthesis of cofactors, prosthetic groups, and carriers Menaquinone and ubiquinone 2-polyprenylphenol 6-hydroxylase (TIGR01982; EC 1.14.13.-; HMM-score: 16.9)early transcribed membrane protein (ETRAMP) family (TIGR01495; HMM-score: 14.5)
- TheSEED :
- Serine/threonine protein kinase PrkC, regulator of stationary phase
A Gram-positive cluster that relates ribosomal protein L28P to a set of uncharacterized proteins Serine/threonine protein kinase PrkC, regulator of stationary phaseand 1 more - PFAM: PKinase (CL0016) Pkinase; Protein kinase domain (PF00069; HMM-score: 199.9)and 11 moreno clan defined PASTA; PASTA domain (PF03793; HMM-score: 158.9)PKinase (CL0016) PK_Tyr_Ser-Thr; Protein tyrosine and serine/threonine kinase (PF07714; HMM-score: 134.4)E-set (CL0159) PrkC-like_PASTA-like; Serine/threonine protein kinase-like, C-terminal PASTA-like domain (PF21160; HMM-score: 133.3)PKinase (CL0016) ABC1; ABC1 atypical kinase-like domain (PF03109; HMM-score: 25.6)APH; Phosphotransferase enzyme family (PF01636; HMM-score: 20)RIO1; RIO domain (PF01163; HMM-score: 16.6)no clan defined Sigma_reg_N; Sigma factor regulator N-terminal (PF13800; HMM-score: 16)PKinase (CL0016) Pkinase_fungal; Fungal protein kinase (PF17667; HMM-score: 16)Kinase-like; Kinase-like (PF14531; HMM-score: 15.7)no clan defined ETRAMP; Malarial early transcribed membrane protein (ETRAMP) (PF09716; HMM-score: 14.1)PKinase (CL0016) TCAD9; Ternary complex associated domain 9 (PF19974; HMM-score: 9.1)
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors:
- effectors:
⊟Localization[edit | edit source]
- PSORTb: Cytoplasmic Membrane
- Cytoplasmic Score: 0.17
- Cytoplasmic Membrane Score: 9.51
- Cellwall Score: 0.16
- Extracellular Score: 0.15
- Internal Helix: 1
- DeepLocPro: Cytoplasmic Membrane
- Cytoplasmic Score: 0
- Cytoplasmic Membrane Score: 0.9728
- Cell wall & surface Score: 0.0001
- Extracellular Score: 0.027
- LocateP: Intracellular /TMH start AFTER 60
- Prediction by SwissProt Classification: Cytoplasmic
- Pathway Prediction: Possibly Sec-
- Intracellular possibility: 0.17
- Signal peptide possibility: -1
- N-terminally Anchored Score: 1
- Predicted Cleavage Site: No CleavageSite
- SignalP: no predicted signal peptide
- SP(Sec/SPI): 0.002877
- TAT(Tat/SPI): 0.000159
- LIPO(Sec/SPII): 0.00064
- predicted transmembrane helices (TMHMM): 1
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MIGKIINERYKIVDKLGGGGMSTVYLAEDTILNIKVAIKAIFIPPREKEETLKRFEREVHNSSQLSHQNIVSMIDVDEEDDCYYLVMEYIEGPTLSEYIESHGPLSVDTAINFTNQILDGIKHAHDMRIVHRDIKPQNILIDSNKTLKIFDFGIAKALSETSLTQTNHVLGTVQYFSPEQAKGEATDECTDIYSIGIVLYEMLVGEPPFNGETAVSIAIKHIQDSVPNVTTDVRKDIPQSLSNVILRATEKDKANRYKTIQEMKDDLSSVLHENRANEDVYELDKMKTIAVPLKKEDLAKHISEHKSNQPKRETTQVPIVNGPAHHQQFQKPEGTVYEPKPKKKSTRKIVLLSLIFSLLMIALVSFVAMAMFGNKYEETPDVIGKSVKEAEQIFNKNNLKLGKISRSYSDKYPENEIIKTTPNTGERVERGDSVDVVISKGPEKVKMPNVIGLPKEEALQKLKSLGLKDVTIEKVYNNQAPKGYIANQSVTANTEIAIHDSNIKLYESLGIKQVYVEDFEHKSFSKAKKALEEKGFKVESKEEYSDDIDEGDVISQSPKGKSVDEGSTISFVVSKGKKSDSSDVKTTTESVDVPYTGKNDKSQKVKVYIKDKDNDGSTEKGSFDITSDQRIDIPLRIEKGKTASYIVKVDGKTVAEKEVSYDDV
⊟Experimental data[edit | edit source]
- experimentally validated: PeptideAtlas [2] [3]
- protein localization:
- quantitative data / protein copy number per cell:
- interaction partners:
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
- MicrobesOnline: SAOUHSC_01182 > SAOUHSC_01183 > SAOUHSC_01184 > SAOUHSC_01185 > SAOUHSC_01186 > SAOUHSC_01187predicted SigA promoter [4] : SAOUHSC_01182 > SAOUHSC_01183 > SAOUHSC_01184 > SAOUHSC_01185 > SAOUHSC_01186 > SAOUHSC_01187
⊟Regulation[edit | edit source]
- regulator:
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: [4]
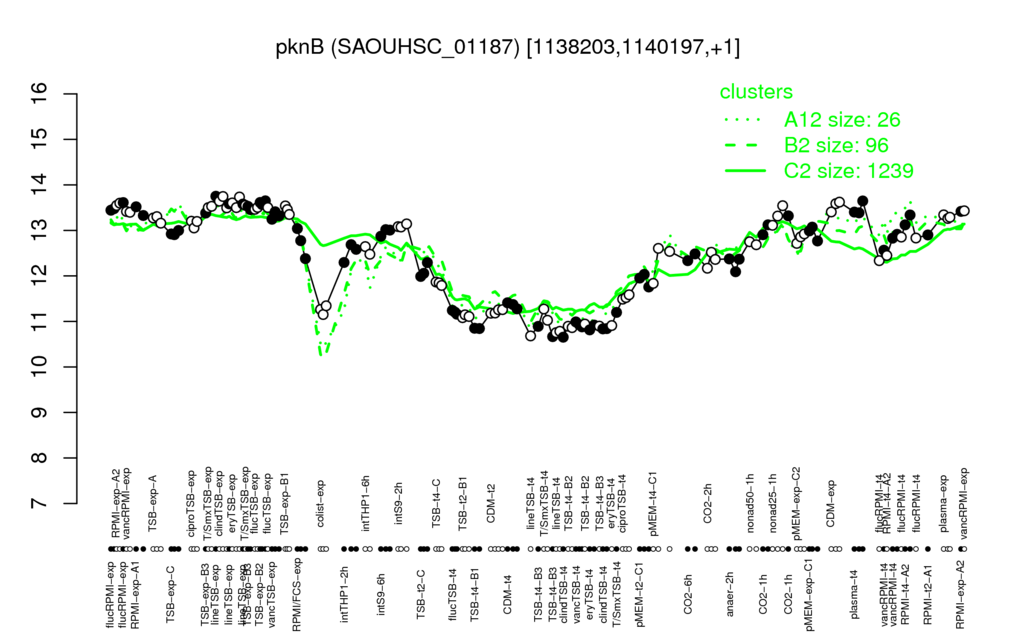 Multi-gene expression profiles
Multi-gene expression profiles
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
- Aureolib: no data available
⊟Protein stability[edit | edit source]
- half-life: no data available
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You can add further information about the gene and protein here. [edit]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ Roy R Chaudhuri, Andrew G Allen, Paul J Owen, Gil Shalom, Karl Stone, Marcus Harrison, Timothy A Burgis, Michael Lockyer, Jorge Garcia-Lara, Simon J Foster, Stephen J Pleasance, Sarah E Peters, Duncan J Maskell, Ian G Charles
Comprehensive identification of essential Staphylococcus aureus genes using Transposon-Mediated Differential Hybridisation (TMDH).
BMC Genomics: 2009, 10;291
[PubMed:19570206] [WorldCat.org] [DOI] (I e) - ↑ Maren Depke, Stephan Michalik, Alexander Rabe, Kristin Surmann, Lars Brinkmann, Nico Jehmlich, Jörg Bernhardt, Michael Hecker, Bernd Wollscheid, Zhi Sun, Robert L Moritz, Uwe Völker, Frank Schmidt
A peptide resource for the analysis of Staphylococcus aureus in host-pathogen interaction studies.
Proteomics: 2015, 15(21);3648-61
[PubMed:26224020] [WorldCat.org] [DOI] (I p) - ↑ Stephan Michalik, Maren Depke, Annette Murr, Manuela Gesell Salazar, Ulrike Kusebauch, Zhi Sun, Tanja C Meyer, Kristin Surmann, Henrike Pförtner, Petra Hildebrandt, Stefan Weiss, Laura Marcela Palma Medina, Melanie Gutjahr, Elke Hammer, Dörte Becher, Thomas Pribyl, Sven Hammerschmidt, Eric W Deutsch, Samuel L Bader, Michael Hecker, Robert L Moritz, Ulrike Mäder, Uwe Völker, Frank Schmidt
A global Staphylococcus aureus proteome resource applied to the in vivo characterization of host-pathogen interactions.
Sci Rep: 2017, 7(1);9718
[PubMed:28887440] [WorldCat.org] [DOI] (I e) - ↑ 4.0 4.1 Ulrike Mäder, Pierre Nicolas, Maren Depke, Jan Pané-Farré, Michel Debarbouille, Magdalena M van der Kooi-Pol, Cyprien Guérin, Sandra Dérozier, Aurelia Hiron, Hanne Jarmer, Aurélie Leduc, Stephan Michalik, Ewoud Reilman, Marc Schaffer, Frank Schmidt, Philippe Bessières, Philippe Noirot, Michael Hecker, Tarek Msadek, Uwe Völker, Jan Maarten van Dijl
Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions.
PLoS Genet: 2016, 12(4);e1005962
[PubMed:27035918] [WorldCat.org] [DOI] (I e)
⊟Relevant publications[edit | edit source]
Stefanie Donat, Karin Streker, Tanja Schirmeister, Sonja Rakette, Thilo Stehle, Manuel Liebeke, Michael Lalk, Knut Ohlsen
Transcriptome and functional analysis of the eukaryotic-type serine/threonine kinase PknB in Staphylococcus aureus.
J Bacteriol: 2009, 191(13);4056-69
[PubMed:19376851] [WorldCat.org] [DOI] (I p)Malgorzata Miller, Stefanie Donat, Sonja Rakette, Thilo Stehle, Thijs R H M Kouwen, Sander H Diks, Annette Dreisbach, Ewoud Reilman, Katrin Gronau, Dörte Becher, Maikel P Peppelenbosch, Jan Maarten van Dijl, Knut Ohlsen
Staphylococcal PknB as the first prokaryotic representative of the proline-directed kinases.
PLoS One: 2010, 5(2);e9057
[PubMed:20140229] [WorldCat.org] [DOI] (I e)
