Jump to navigation
Jump to search
NCBI: 03-AUG-2016
⊟Summary[edit | edit source]
- organism: Staphylococcus aureus NCTC8325
- locus tag: SAOUHSC_01974
- pan locus tag?: SAUPAN004773000
- symbol: SAOUHSC_01974
- pan gene symbol?: —
- synonym:
- product: hypothetical protein
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
- type: CDS
- locus tag: SAOUHSC_01974
- symbol: SAOUHSC_01974
- product: hypothetical protein
- replicon: chromosome
- strand: -
- coordinates: 1877270..1880206
- length: 2937
- essential: no DEG other strains
⊟Accession numbers[edit | edit source]
- Gene ID: 3920450 NCBI
- RefSeq: YP_500471 NCBI
- BioCyc: G1I0R-1840 BioCyc
- MicrobesOnline: 1290393 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421
481
541
601
661
721
781
841
901
961
1021
1081
1141
1201
1261
1321
1381
1441
1501
1561
1621
1681
1741
1801
1861
1921
1981
2041
2101
2161
2221
2281
2341
2401
2461
2521
2581
2641
2701
2761
2821
2881ATGATAATTAAATCACTTGAAATTTATGGTTACGGTCAATTTGTTCAACGTAAAATTGAA
TTTAATAAAAACTTCACTGAAATTTTTGGTGAAAATGAAGCGGGTAAATCGACGATTCAA
GCATTCATCCATTCGATATTATTTGGATTTCCAACTAAAAAGTCTAAAGAGCCAAGACTA
GAACCACGTCTAGGTAACCAATACGGTGGTAAATTAGTACTTATTCTTGATGATGGCTTA
GAGATTGAAGTTGAACGAATTAAAGGCAGTGCTCAAGGTGATGTGAAAGTATATTTACCT
AATGGTGCTGTGCGTGATGATGCTTGGTTACAAAAGAAACTTAATTATATTTCTAAAAAG
ACATATCAAGGTATCTTTTCATTTGATGTACTAGGGCTTCAAGACATTCATAGAAATCTA
AATGAAAAACAATTGCAAGATTATTTATTACAAGCAGGGGCTTTAGGATCAACTGAATTC
ACGTCAATGCGCGAAGTGATTAATCGTAAAAAAGATGAATTATATAAAAAATCAGGTAAA
AATCCGATCATTAATCAACAAATTGAGCAATTAAAACAACTAGAAAGTCAAATTCGTGAA
GAAGAAGCAAAGCTAGAAACATATCATCGCTTAGTAGATGATCGAGATAAATCATCACGT
CGATTAGAGAATTTAAAGCATAATTTAAATCAATTATCAAAAATGCATGAAGAAAAACAA
AAAGAGGTTGCTTTACATGATCATTCACAAGAATGGAAGTCTCTAGAACAACAGTTAAAT
ATTGAGCCAATCACATTCCCAGAAAAAGGTGTGGATCGTTACGAAAAAGCACGAGCGCAT
AAGCAATCGTTAGAAAGAGATATTGGTTTAAGAAATGAGCGTTTAGCTCAACTTAAAGAA
GAAGCGACTCAATTAGAGCCAGTTAAACAATCTGATATTGACGCCTTCATTAGTTTGAAT
CAACAAGAAAATGAAATTAAAAATAAAGAATTTGAACTTACTGCAATCGAAAAGGATATT
GCGAATAAACAACGTGATAAAGATGAATTGCAATCAAATATTGGTTGGTCTGAAACGCAT
CATGACGTAGATAGTTCAGAGGCAATGAAAAGTTATGTCAGTGAGCAAATCAAGAATAAA
CAAGAACAAGCTGCATACATTAAACAATTAGAACGTAGTTTAGAAGAAAATAAAATCGAA
GATAATGCGGTTCATAGCGAACTAGATTCTGTTGAAGAAAAAATAGTTCCTGAAGAGACT
TTTGAAAAGAAAAAAGAATACTCACAACAAGTCATTGAATTAAACGAAAAAGAGAACTTG
TATAGCAAATTGAAAGAACGTTTCGAAATTGAGCAACAAGAAAAACAAAAACGTCAAAAA
TTGTTACGAACAACGTTTATTCTTTTAACATTAGTAGGTATTGGTTTAACTGCCTTCTCA
TTTATTTCAAACAACATGTTGTTTGGTATTATTTTCGCAGTATTAACTTTAGTATTTGTT
ATCGGTATCATCATGTCTAAATCAAAAGAAGTAGATTATAGCGAGGCCATCACAGATGAA
ATCGAAGAAATTAAAGCACAACTAGCCATTTTAGATGAAAATTATGACTTAGATTTTGAT
CTTGATGAGCAATATCGCATTCGTGATCACTGGCAACAAGCTTTGAAAAATAAAGATATT
TTAGAAGAGAAACGTCAGTATATTGAAGGTCGTTTAAATGATGCGAAAGGGCGTCATGAT
GAATTACAATCGACAGTTGAGAACGTTAAGGATGAATTATACTTATCATCTAAAATTTCA
AATGATTTAATTGTTGATAGTATTTCTACAATGGCGAATATTAAAGCATTAGATCAACAT
ATCAGTGATTTAAATCAACAACGCCAGCAATTGGTTCAAGAATTAGATACATTCTACAAT
CATGCTGAAGCTGTTACAAAATCACAATTTGTTTATTTCAACAAATTATCTTTATTCCAC
GATGTGCAACAATGGTTGAAATCAGCTGAAGACACGAATGAAAAATGGCGTATTAATGCT
GAAAATACCAAACTTGTTACAAATGAATTAAATCATTTAAATGCTCAATTAGAAGAAAAT
AATAAAGAAATTACAGCACTATTCGATTTTATCAATGTTGGTACTGAAGAAGATTTTTAC
CAACATCATGAAGATTACCAAACTTATACTAGTAATTTGAGTCGTTTTAATGATTTAACT
AAATATCTTGAGAACCAAAACTATTCTTATGAATTAAGTTCTAGTTTAAGTGAAAAAACA
ACTGCACAACTTGAAGAAGAAGATCATTTATTGGCTACTCAAGTTGACGAATATAATGAG
CAATATCTTGAAATGCAAGCACAAGTCAGTGATTTAAGTGCACAAATCAATCACATGGAA
ACTGATACAACGCTTGCTAATTTAAGACATGAATATCACAGTCTTAAAAATCAACTTAAT
GATATCGCAAAAGATTGGGCAAGTTTAAGTTATTTACAAAGTTTAGTTGATGAACACATT
AAACAAATTAAAGATAAACGTTTGCCTCAAGTTATTAATGAAGCGGTAGAAATATTGAAG
CATTTAACAGATGGCAGATATACGATGATTAACTATAATGAAGATTCAATTACGGTTAAA
CATGTTAATGGTCAATTATATGATCCTGTTGAACTAAGTCAATCTACAAAAGAATTACTT
TATGTAGCTTTACGTATCAGTTTAATTAAGGTACTAAGACCATATTATCCGTTCCCATTA
ATTGTTGATGATGCATTTGTTCATTTTGATAAAAAACGTACTGAAAAAATGTTGAATTAT
TTACGATCATTATCAGAACACTATCAAGTACTATACTTTACATGTGTAAAAGATAATATT
GTTCCATCAAAAGAAGTGATTACATTAAACAAAATAGAGGAAGGCGGGAAACGATGA60
120
180
240
300
360
420
480
540
600
660
720
780
840
900
960
1020
1080
1140
1200
1260
1320
1380
1440
1500
1560
1620
1680
1740
1800
1860
1920
1980
2040
2100
2160
2220
2280
2340
2400
2460
2520
2580
2640
2700
2760
2820
2880
2937
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SAOUHSC_01974
- symbol: SAOUHSC_01974
- description: hypothetical protein
- length: 978
- theoretical pI: 4.97145
- theoretical MW: 114420
- GRAVY: -0.726278
⊟Function[edit | edit source]
- TIGRFAM: Cellular processes Cell division chromosome segregation protein SMC (TIGR02169; HMM-score: 17.7)DNA metabolism Chromosome-associated proteins chromosome segregation protein SMC (TIGR02169; HMM-score: 17.7)Cellular processes Cell division chromosome segregation protein SMC (TIGR02168; HMM-score: 17.6)DNA metabolism Chromosome-associated proteins chromosome segregation protein SMC (TIGR02168; HMM-score: 17.6)
- TheSEED :
- DNA double-strand break repair rad50 ATPase
- PFAM: P-loop_NTPase (CL0023) AAA_27; AAA domain (PF13514; HMM-score: 89)and 11 moreno clan defined CENP-F_leu_zip; Leucine-rich repeats of kinetochore protein Cenp-F/LEK1 (PF10473; HMM-score: 24.5)Hypoth_1 (CL0447) DUF3784; Domain of unknown function (DUF3784) (PF12650; HMM-score: 19.7)P-loop_NTPase (CL0023) AAA_29; P-loop containing region of AAA domain (PF13555; HMM-score: 18.2)AAA_23; AAA domain (PF13476; HMM-score: 17.3)SMC_N; RecF/RecN/SMC N terminal domain (PF02463; HMM-score: 15.9)SbcC_Walker_B; SbcC/RAD50-like, Walker B motif (PF13558; HMM-score: 15.7)no clan defined DUF4191; Domain of unknown function (DUF4191) (PF13829; HMM-score: 15.1)DUF6249; Domain of unknown function (DUF6249) (PF19762; HMM-score: 14)GPCR_A (CL0192) 7tm_2; 7 transmembrane receptor (Secretin family) (PF00002; HMM-score: 12.2)no clan defined EcsB; Bacterial ABC transporter protein EcsB (PF05975; HMM-score: 12.2)FtsH_ext; FtsH Extracellular (PF06480; HMM-score: 12.1)
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors:
- effectors:
⊟Localization[edit | edit source]
- PSORTb: Cytoplasmic
- Cytoplasmic Score: 7.5
- Cytoplasmic Membrane Score: 1.15
- Cellwall Score: 0.62
- Extracellular Score: 0.73
- Internal Helices: 2
- DeepLocPro: Cytoplasmic Membrane
- Cytoplasmic Score: 0.0189
- Cytoplasmic Membrane Score: 0.9012
- Cell wall & surface Score: 0.0089
- Extracellular Score: 0.071
- LocateP: Multi-transmembrane
- Prediction by SwissProt Classification: Membrane
- Pathway Prediction: Sec-(SPI)
- Intracellular possibility: 0.17
- Signal peptide possibility: -1
- N-terminally Anchored Score: -1
- Predicted Cleavage Site: No CleavageSite
- SignalP: no predicted signal peptide
- SP(Sec/SPI): 0.006246
- TAT(Tat/SPI): 0.000529
- LIPO(Sec/SPII): 0.001162
- predicted transmembrane helices (TMHMM): 2
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MIIKSLEIYGYGQFVQRKIEFNKNFTEIFGENEAGKSTIQAFIHSILFGFPTKKSKEPRLEPRLGNQYGGKLVLILDDGLEIEVERIKGSAQGDVKVYLPNGAVRDDAWLQKKLNYISKKTYQGIFSFDVLGLQDIHRNLNEKQLQDYLLQAGALGSTEFTSMREVINRKKDELYKKSGKNPIINQQIEQLKQLESQIREEEAKLETYHRLVDDRDKSSRRLENLKHNLNQLSKMHEEKQKEVALHDHSQEWKSLEQQLNIEPITFPEKGVDRYEKARAHKQSLERDIGLRNERLAQLKEEATQLEPVKQSDIDAFISLNQQENEIKNKEFELTAIEKDIANKQRDKDELQSNIGWSETHHDVDSSEAMKSYVSEQIKNKQEQAAYIKQLERSLEENKIEDNAVHSELDSVEEKIVPEETFEKKKEYSQQVIELNEKENLYSKLKERFEIEQQEKQKRQKLLRTTFILLTLVGIGLTAFSFISNNMLFGIIFAVLTLVFVIGIIMSKSKEVDYSEAITDEIEEIKAQLAILDENYDLDFDLDEQYRIRDHWQQALKNKDILEEKRQYIEGRLNDAKGRHDELQSTVENVKDELYLSSKISNDLIVDSISTMANIKALDQHISDLNQQRQQLVQELDTFYNHAEAVTKSQFVYFNKLSLFHDVQQWLKSAEDTNEKWRINAENTKLVTNELNHLNAQLEENNKEITALFDFINVGTEEDFYQHHEDYQTYTSNLSRFNDLTKYLENQNYSYELSSSLSEKTTAQLEEEDHLLATQVDEYNEQYLEMQAQVSDLSAQINHMETDTTLANLRHEYHSLKNQLNDIAKDWASLSYLQSLVDEHIKQIKDKRLPQVINEAVEILKHLTDGRYTMINYNEDSITVKHVNGQLYDPVELSQSTKELLYVALRISLIKVLRPYYPFPLIVDDAFVHFDKKRTEKMLNYLRSLSEHYQVLYFTCVKDNIVPSKEVITLNKIEEGGKR
⊟Experimental data[edit | edit source]
- experimentally validated: PeptideAtlas [1] [2]
- protein localization: data available for COL
- quantitative data / protein copy number per cell:
- interaction partners:
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
⊟Regulation[edit | edit source]
- regulator:
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: [3]
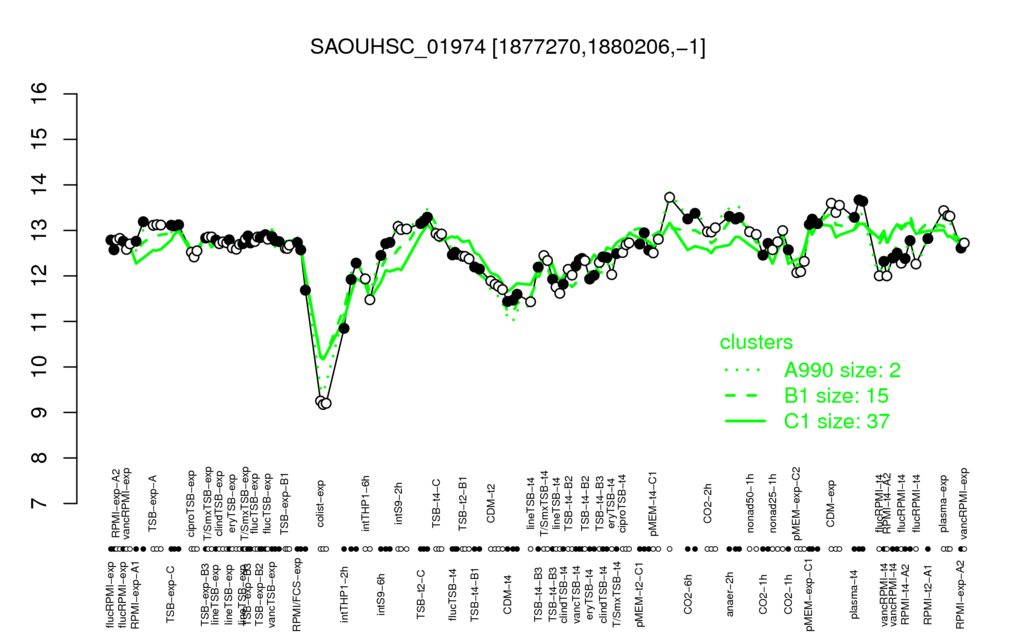 Multi-gene expression profiles
Multi-gene expression profiles
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
- Aureolib: no data available
⊟Protein stability[edit | edit source]
- half-life: no data available
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You can add further information about the gene and protein here. [edit]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ Maren Depke, Stephan Michalik, Alexander Rabe, Kristin Surmann, Lars Brinkmann, Nico Jehmlich, Jörg Bernhardt, Michael Hecker, Bernd Wollscheid, Zhi Sun, Robert L Moritz, Uwe Völker, Frank Schmidt
A peptide resource for the analysis of Staphylococcus aureus in host-pathogen interaction studies.
Proteomics: 2015, 15(21);3648-61
[PubMed:26224020] [WorldCat.org] [DOI] (I p) - ↑ Stephan Michalik, Maren Depke, Annette Murr, Manuela Gesell Salazar, Ulrike Kusebauch, Zhi Sun, Tanja C Meyer, Kristin Surmann, Henrike Pförtner, Petra Hildebrandt, Stefan Weiss, Laura Marcela Palma Medina, Melanie Gutjahr, Elke Hammer, Dörte Becher, Thomas Pribyl, Sven Hammerschmidt, Eric W Deutsch, Samuel L Bader, Michael Hecker, Robert L Moritz, Ulrike Mäder, Uwe Völker, Frank Schmidt
A global Staphylococcus aureus proteome resource applied to the in vivo characterization of host-pathogen interactions.
Sci Rep: 2017, 7(1);9718
[PubMed:28887440] [WorldCat.org] [DOI] (I e) - ↑ 3.0 3.1 Ulrike Mäder, Pierre Nicolas, Maren Depke, Jan Pané-Farré, Michel Debarbouille, Magdalena M van der Kooi-Pol, Cyprien Guérin, Sandra Dérozier, Aurelia Hiron, Hanne Jarmer, Aurélie Leduc, Stephan Michalik, Ewoud Reilman, Marc Schaffer, Frank Schmidt, Philippe Bessières, Philippe Noirot, Michael Hecker, Tarek Msadek, Uwe Völker, Jan Maarten van Dijl
Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions.
PLoS Genet: 2016, 12(4);e1005962
[PubMed:27035918] [WorldCat.org] [DOI] (I e)
