Jump to navigation
Jump to search
NCBI: 03-AUG-2016
⊟Summary[edit | edit source]
- organism: Staphylococcus aureus NCTC8325
- locus tag: SAOUHSC_02681
- pan locus tag?: SAUPAN005928000
- symbol: SAOUHSC_02681
- pan gene symbol?: narG
- synonym:
- product: nitrate reductase subunit alpha
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
- type: CDS
- locus tag: SAOUHSC_02681
- symbol: SAOUHSC_02681
- product: nitrate reductase subunit alpha
- replicon: chromosome
- strand: -
- coordinates: 2463891..2467580
- length: 3690
- essential: no DEG other strains
⊟Accession numbers[edit | edit source]
- Gene ID: 3921243 NCBI
- RefSeq: YP_501143 NCBI
- BioCyc: G1I0R-2527 BioCyc
- MicrobesOnline: 1291114 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421
481
541
601
661
721
781
841
901
961
1021
1081
1141
1201
1261
1321
1381
1441
1501
1561
1621
1681
1741
1801
1861
1921
1981
2041
2101
2161
2221
2281
2341
2401
2461
2521
2581
2641
2701
2761
2821
2881
2941
3001
3061
3121
3181
3241
3301
3361
3421
3481
3541
3601
3661ATGGGAAAATTTGGATTGAATTTCTTTAAGCCAACAGAAAAATTTAATGGGAATTGGTCG
ATCCTAGAAAGTAAAAGTAGAGAATGGGAAAAAATGTACAGAGAACGTTGGAGCCACGAT
AAAGAAGTAAGAACAACACATGGTGTTAACTGTACAGGCTCATGTTCTTGGAAAGTATTT
GTGAAAAATGGTGTGATTACCTGGGAAAATCAACAAACTGACTATCCAAGTTGTGGTCCG
GATATGCCTGAATATGAACCGAGAGGATGTCCACGAGGTGCGTCATTCTCTTGGTATGAA
TACAGTCCGCTTCGAATCAAATATCCATATATTCGTGGAAAACTCTGGGATTTATGGACT
GAAGCATTAGAAGAAAACAATGGTAATCGCGTTGCTGCATGGGCGTCTATTGTTGAAAAT
GAAGACAAAGCCAAACAATATAAGCAAGCCCGAGGTATGGGAGGGCACGTGCGTTCAAAT
TGGAAAGACGTTACAGAGATAATCGCAGCACAATTACTGTATACAATAAAAAAATATGGT
CCAGATCGAATCGCAGGATTTACACCTATTCCAGCGATGTCAATGATTAGTTATGCAGCA
GGTGCTCGATTCATCAATTTGCTTGGTGGTGAAATGCTTAGTTTTTATGACTGGTATGCA
GATTTACCACCTGCCTCTCCACAAATTTGGGGAGAGCAAACAGATGTGCCTGAATCAAGT
GACTGGTATAACGCATCATACATTATTATGTGGGGCTCTAATGTACCTTTAACACGTACT
CCGGATGCACATTTTATGACTGAAGTCCGCTATAAAGGTACAAAAGTCATTTCAGTAGCA
CCAGATTACGCAGAAAATGTGAAATTTGCAGATAACTGGCTAGCACCGAATCCTGGTTCA
GATGCTGCAATTGCACAAGCAATGACGCATGTTATTTTACAAGAACATTATGTTAATCAA
CCTAATGAACGCTTTATAAATTACGCTAAACAATATACAGATATGCCGTTTCTTATCATG
CTGGATGAAGATGAAAATGGATATAAAGCGGGTCGATTTTTAAGAGCGAGTGACTTAGGT
CAAACAACAGAGCAAGGCGAATGGAAGCCAGTTATTCATGATGCAATCAGCGATAGTTTA
GTAGTACCTAATGGCACAATGGGTCAACGTTGGGAAGAAGGTAAGAAGTGGAACTTAAAA
CTAGAAACAGAAGATGGTTCTAAAATTAACCCTACATTATCAATGACAGAAGGTGGATAC
GAATTAGAAACAATTCAATTCCCATACTTTGATAGTGATGGAGATGGGATATTCAATCGT
CCAATTCCAACTCGACAAGTCACTTTAGCAAATGGTGACAAAGTCCGTATTGCTACAATT
TTTGACTTAATGGCAAGTCAATATGGCGTGCGTCGTTTTGATCATAAATTAGAATCAAAA
GGATACGACGATGCAGAATCAAAATATACACCTGCTTGGCAAGAAGCCATTTCAGGCGTA
AAACAAAGTGTTGTCATTCAAGTAGCGAAAGAATTTGCGCAAAACGCTATCGATACTGAA
GGGCGTTCAATGATTATCATGGGTGCGGGTATTAACCATTGGTTTAACTCAGATACGATT
TATCGTTCAATCTTAAACTTAGTTATGTTATGTGGCTGTCAAGGTGTGAATGGTGGCGGT
TGGGCTCACTATGTGGGACAAGAAAAATGTCGTCCGATTGAAGGATGGAGTACTGTCGCA
TTTGCGAAAGACTGGCAAGGACCACCACGTTTGCAAAACGGAACAAGTTGGTTCTATTTT
GCAACAGACCAATGGAAATATGAAGAGTCAAATGTAGATCGTTTAAAATCTCCATTAGCT
AAAACAGAGGAGTTAAAGCATCAACATCCAGCTGATTATAATGTTTTAGCAGCTAGACTT
GGTTGGTTACCATCATATCCACAATTTAATAAAAATAGTTTGTTGTTTGCAGAAGAAGCT
AAAGATGAAGGTATAGATTCAAATGAAGCGATTTTGCAACGTGCAATTGATGAAGTGAAA
TCAAAACAAACACAATTTGCAATAGAAGATCCTGATTTGAAAAAGAATCATCCAAAATCA
TTATTTATATGGCGTTCAAACTTAATTTCAAGTTCTGCAAAAGGTCAAGAATACTTTATG
AAGCATTTACTTGGCACAAAATCAGGGTTATTAGCTACACCAAATGAAGATGAAAAGCCA
GAAGAAATTACGTGGCGTGAAGAAACAACAGGTAAGTTAGATTTAGTGGTCTCTTTAGAC
TTCAGAATGACGGCAACACCGTTATATTCTGACATTGTTTTGCCGGCAGCGACTTGGTAT
GAAAAACATGATTTATCATCAACGGATATGCATCCATATGTACATCCTTTCAATCCTGCC
ATTGATCCGTTATGGGAATCGCGTTCAGACTGGGATATTTATAAAACGTTGGCAAAAGCA
TTTTCAGAAATGGCAAAAGACTATTTACCTGGAACGTTTAAAGATGTTGTGACAACTCCA
CTTAGTCATGATACAAAGCAAGAAATTTCAACACCATACGGCGTAGTGAAAGATTGGTCG
AAGGGTGAAATTGAAGCGGTACCTGGACGTACAATGCCTAACTTTGCAATTGTAGAACGC
GACTACACTAAAATTTACGACAAATATGTCACGCTTGGTCCTGTACTTGAAAAAGGGAAA
GTTGGAGCACATGGTGTAAGTTTCGGTGTCAGTGAACAATATGAAGAATTAAAAAGTATG
TTAGGTACGTGGAGTGATACAAATGATGATTCTGTGAGAGCGAATCGTCCACGTATTGAT
ACAGCACGTAATGTAGCTGATGCAATACTAAGTATTTCATCTGCTACGAATGGTAAATTA
TCACAAAAATCATATGAAGATCTCGAAGAACAAACTGGAATGCCGTTAAAAGATATTTCT
AGCGAACGCGCTGCTGAGAAAATTTCGTTTTTAAATATAACTTCACAACCACGAGAAGTA
ATACCGACAGCAGTATTCCCAGGTTCAAATAAACAAGGTCGACGATATTCACCATTTACA
ACGAATATAGAACGTCTAGTACCTTTTAGAACATTAACAGGACGTCAAAGTTATTATGTG
GATCACGAAGTTTTCCAACAATTTGGGGAGAGCTTACCAGTATATAAACCGACATTGCCG
CCAATGGTATTTGGGAATAGAGATAAGAAAATTAAAGGTGGTACAGATGCTTTGGTACTG
CGTTATTTAACGCCTCATGGAAAATGGAATATACACTCAATGTATCAAGATAATAAGCAT
ATGTTGACACTATTTAGAGGTGGTCCAACGGTTTGGATATCAAATGAAGATGCTGAAAAA
CACGATATCCAAGATAATGATTGGCTAGAAGTGTATAACCGTAATGGTGTTGTAACGGCA
AGAGCAGTTATTTCGCATCGTATGCCTAAAGGTACAATGTTTATGTATCATGCACAAGAT
AAACATATTCAAACGCCTGGGTCAGAAATTACAGATACACGTGGTGGTTCACACAACGCG
CCGACTAGAATCCATTTGAAACCAACACAACTAGTCGGAGGATACGCACAAATTAGTTAT
CACTTTAATTATTATGGACCAATTGGGAACCAAAGGGATTTATATGTAGCAGTTAGAAAG
ATGAAGGAGGTTAATTGGCTTGAAGATTAA60
120
180
240
300
360
420
480
540
600
660
720
780
840
900
960
1020
1080
1140
1200
1260
1320
1380
1440
1500
1560
1620
1680
1740
1800
1860
1920
1980
2040
2100
2160
2220
2280
2340
2400
2460
2520
2580
2640
2700
2760
2820
2880
2940
3000
3060
3120
3180
3240
3300
3360
3420
3480
3540
3600
3660
3690
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SAOUHSC_02681
- symbol: SAOUHSC_02681
- description: nitrate reductase subunit alpha
- length: 1229
- theoretical pI: 6.19853
- theoretical MW: 139828
- GRAVY: -0.592596
⊟Function[edit | edit source]
- reaction: EC 1.7.99.4? ExPASyNitrate reductase Nitrite + acceptor = nitrate + reduced acceptor
- TIGRFAM: Energy metabolism Anaerobic nitrate reductase, alpha subunit (TIGR01580; EC 1.7.99.4; HMM-score: 1838.2)and 13 moreDMSO reductase family type II enzyme, molybdopterin subunit (TIGR03479; HMM-score: 591.5)anaerobic dimethyl sulfoxide reductase, A subunit, DmsA/YnfE family (TIGR02166; HMM-score: 180.3)formate dehydrogenase, alpha subunit (TIGR01591; EC 1.2.1.2; HMM-score: 132.8)molybdopterin guanine dinucleotide-containing S/N-oxide reductases (TIGR00509; HMM-score: 102.5)trimethylamine-N-oxide reductase TorA (TIGR02164; EC 1.7.2.3; HMM-score: 78.6)Energy metabolism Anaerobic formate dehydrogenase-N alpha subunit (TIGR01553; EC 1.1.5.6; HMM-score: 46.7)Energy metabolism Electron transport formate dehydrogenase-N alpha subunit (TIGR01553; EC 1.1.5.6; HMM-score: 46.7)Energy metabolism Electron transport arsenite oxidase, large subunit (TIGR02693; EC 1.20.9.1; HMM-score: 28.3)Central intermediary metabolism Nitrogen metabolism periplasmic nitrate reductase, large subunit (TIGR01706; EC 1.7.99.4; HMM-score: 24.6)Energy metabolism Electron transport periplasmic nitrate reductase, large subunit (TIGR01706; EC 1.7.99.4; HMM-score: 24.6)Energy metabolism Aerobic periplasmic nitrate reductase, large subunit (TIGR01706; EC 1.7.99.4; HMM-score: 24.6)oxidoreductase alpha (molybdopterin) subunit (TIGR01701; HMM-score: 19.7)Energy metabolism Electron transport NADH dehydrogenase (quinone), G subunit (TIGR01973; EC 1.6.99.5; HMM-score: 13.9)
- TheSEED :
- Respiratory nitrate reductase (quinol), alpha subunit (EC 1.7.5.1)
- PFAM: no clan defined Molybdopterin; Molybdopterin oxidoreductase (PF00384; HMM-score: 241.4)and 4 moreDPBB (CL0199) Molydop_binding; Molydopterin dinucleotide binding domain (PF01568; HMM-score: 74.2)4Fe-4S (CL0344) Nitr_red_alph_N; Respiratory nitrate reductase alpha N-terminal (PF14710; HMM-score: 38)Molybdop_Fe4S4; Molybdopterin oxidoreductase Fe4S4 domain (PF04879; HMM-score: 13.7)PUA (CL0178) PUA_3; PUA-like domain (PF17785; HMM-score: 12.2)
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors: Mo-bis(molybdopterin guanine dinucleotide), [4Fe-4S] cluster
- effectors:
⊟Localization[edit | edit source]
- PSORTb: Cytoplasmic Membrane
- Cytoplasmic Score: 0.01
- Cytoplasmic Membrane Score: 9.99
- Cellwall Score: 0
- Extracellular Score: 0
- Internal Helices: 0
- DeepLocPro: Cytoplasmic Membrane
- Cytoplasmic Score: 0.0426
- Cytoplasmic Membrane Score: 0.5363
- Cell wall & surface Score: 0.0003
- Extracellular Score: 0.4208
- LocateP: Intracellular
- Prediction by SwissProt Classification: Cytoplasmic
- Pathway Prediction: No pathway
- Intracellular possibility: 1
- Signal peptide possibility: -1
- N-terminally Anchored Score: 1
- Predicted Cleavage Site: No CleavageSite
- SignalP: no predicted signal peptide
- SP(Sec/SPI): 0.069378
- TAT(Tat/SPI): 0.00154
- LIPO(Sec/SPII): 0.003017
- predicted transmembrane helices (TMHMM): 0
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MGKFGLNFFKPTEKFNGNWSILESKSREWEKMYRERWSHDKEVRTTHGVNCTGSCSWKVFVKNGVITWENQQTDYPSCGPDMPEYEPRGCPRGASFSWYEYSPLRIKYPYIRGKLWDLWTEALEENNGNRVAAWASIVENEDKAKQYKQARGMGGHVRSNWKDVTEIIAAQLLYTIKKYGPDRIAGFTPIPAMSMISYAAGARFINLLGGEMLSFYDWYADLPPASPQIWGEQTDVPESSDWYNASYIIMWGSNVPLTRTPDAHFMTEVRYKGTKVISVAPDYAENVKFADNWLAPNPGSDAAIAQAMTHVILQEHYVNQPNERFINYAKQYTDMPFLIMLDEDENGYKAGRFLRASDLGQTTEQGEWKPVIHDAISDSLVVPNGTMGQRWEEGKKWNLKLETEDGSKINPTLSMTEGGYELETIQFPYFDSDGDGIFNRPIPTRQVTLANGDKVRIATIFDLMASQYGVRRFDHKLESKGYDDAESKYTPAWQEAISGVKQSVVIQVAKEFAQNAIDTEGRSMIIMGAGINHWFNSDTIYRSILNLVMLCGCQGVNGGGWAHYVGQEKCRPIEGWSTVAFAKDWQGPPRLQNGTSWFYFATDQWKYEESNVDRLKSPLAKTEELKHQHPADYNVLAARLGWLPSYPQFNKNSLLFAEEAKDEGIDSNEAILQRAIDEVKSKQTQFAIEDPDLKKNHPKSLFIWRSNLISSSAKGQEYFMKHLLGTKSGLLATPNEDEKPEEITWREETTGKLDLVVSLDFRMTATPLYSDIVLPAATWYEKHDLSSTDMHPYVHPFNPAIDPLWESRSDWDIYKTLAKAFSEMAKDYLPGTFKDVVTTPLSHDTKQEISTPYGVVKDWSKGEIEAVPGRTMPNFAIVERDYTKIYDKYVTLGPVLEKGKVGAHGVSFGVSEQYEELKSMLGTWSDTNDDSVRANRPRIDTARNVADAILSISSATNGKLSQKSYEDLEEQTGMPLKDISSERAAEKISFLNITSQPREVIPTAVFPGSNKQGRRYSPFTTNIERLVPFRTLTGRQSYYVDHEVFQQFGESLPVYKPTLPPMVFGNRDKKIKGGTDALVLRYLTPHGKWNIHSMYQDNKHMLTLFRGGPTVWISNEDAEKHDIQDNDWLEVYNRNGVVTARAVISHRMPKGTMFMYHAQDKHIQTPGSEITDTRGGSHNAPTRIHLKPTQLVGGYAQISYHFNYYGPIGNQRDLYVAVRKMKEVNWLED
⊟Experimental data[edit | edit source]
- experimentally validated: PeptideAtlas [1] [2]
- protein localization: data available for COL
- quantitative data / protein copy number per cell:
- interaction partners:
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
- MicrobesOnline: SAOUHSC_02675 < SAOUHSC_02676 < SAOUHSC_02677 < SAOUHSC_02678 < SAOUHSC_02679 < SAOUHSC_02680 < SAOUHSC_02681predicted SigA promoter [3] : SAOUHSC_02675 < S1044 < SAOUHSC_02676 < SAOUHSC_02677 < SAOUHSC_02678 < SAOUHSC_02679 < SAOUHSC_02680 < S1045 < SAOUHSC_02681 < S1046
⊟Regulation[edit | edit source]
- regulators: Rex* (repression) regulon, NreC* (activation) regulon
Rex* (TF) important in Energy metabolism; RegPrecise NreC* (TF) important in Nitrate and nitrite respiration; RegPrecise
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: [3]
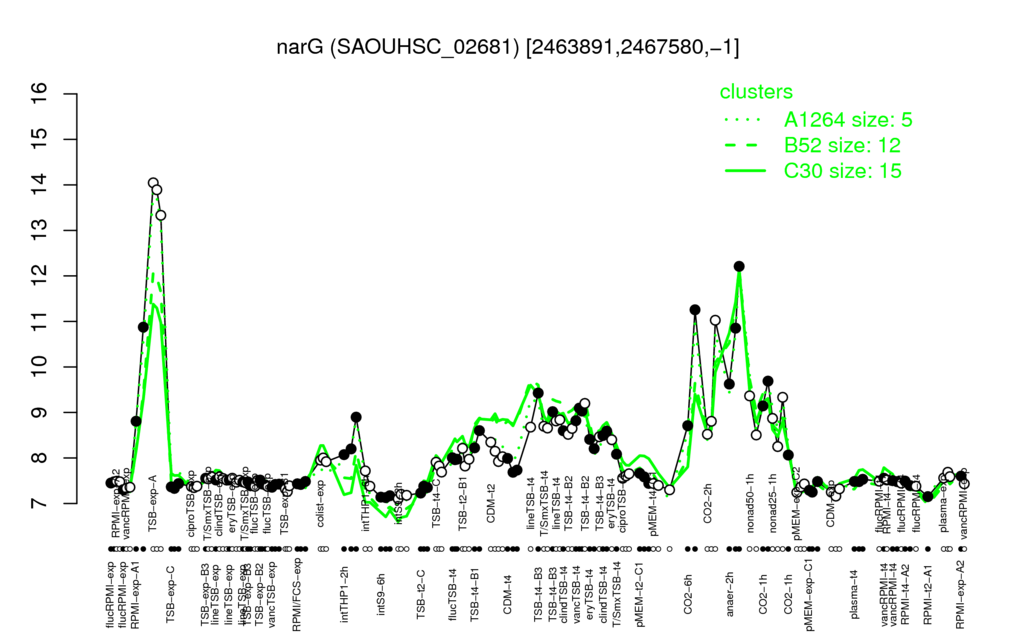 Multi-gene expression profiles
Multi-gene expression profiles
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
- Aureolib: no data available
⊟Protein stability[edit | edit source]
- half-life: no data available
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You can add further information about the gene and protein here. [edit]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ Maren Depke, Stephan Michalik, Alexander Rabe, Kristin Surmann, Lars Brinkmann, Nico Jehmlich, Jörg Bernhardt, Michael Hecker, Bernd Wollscheid, Zhi Sun, Robert L Moritz, Uwe Völker, Frank Schmidt
A peptide resource for the analysis of Staphylococcus aureus in host-pathogen interaction studies.
Proteomics: 2015, 15(21);3648-61
[PubMed:26224020] [WorldCat.org] [DOI] (I p) - ↑ Stephan Michalik, Maren Depke, Annette Murr, Manuela Gesell Salazar, Ulrike Kusebauch, Zhi Sun, Tanja C Meyer, Kristin Surmann, Henrike Pförtner, Petra Hildebrandt, Stefan Weiss, Laura Marcela Palma Medina, Melanie Gutjahr, Elke Hammer, Dörte Becher, Thomas Pribyl, Sven Hammerschmidt, Eric W Deutsch, Samuel L Bader, Michael Hecker, Robert L Moritz, Ulrike Mäder, Uwe Völker, Frank Schmidt
A global Staphylococcus aureus proteome resource applied to the in vivo characterization of host-pathogen interactions.
Sci Rep: 2017, 7(1);9718
[PubMed:28887440] [WorldCat.org] [DOI] (I e) - ↑ 3.0 3.1 Ulrike Mäder, Pierre Nicolas, Maren Depke, Jan Pané-Farré, Michel Debarbouille, Magdalena M van der Kooi-Pol, Cyprien Guérin, Sandra Dérozier, Aurelia Hiron, Hanne Jarmer, Aurélie Leduc, Stephan Michalik, Ewoud Reilman, Marc Schaffer, Frank Schmidt, Philippe Bessières, Philippe Noirot, Michael Hecker, Tarek Msadek, Uwe Völker, Jan Maarten van Dijl
Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions.
PLoS Genet: 2016, 12(4);e1005962
[PubMed:27035918] [WorldCat.org] [DOI] (I e)
