Jump to navigation
Jump to search
m (Text replacement - "gene Genbank" to "gene RefSeq") |
m (Text replacement - "* <aureodatabase>protein Genbank</aureodatabase> " to "") |
||
| Line 1: | Line 1: | ||
__TOC__ | |||
<protect> | <protect> | ||
<aureodatabase> | <aureodatabase>annotation</aureodatabase> | ||
=Summary= | =Summary= | ||
* <aureodatabase>organism</aureodatabase> | *<aureodatabase>organism</aureodatabase> | ||
* <aureodatabase>locus</aureodatabase> | *<aureodatabase>locus</aureodatabase> | ||
* <aureodatabase>pan locus</aureodatabase> | *<aureodatabase>pan locus</aureodatabase> | ||
* <aureodatabase>gene symbol</aureodatabase> | *<aureodatabase>gene symbol</aureodatabase> | ||
* <aureodatabase>pan gene symbol</aureodatabase> | *<aureodatabase>pan gene symbol</aureodatabase> | ||
* <aureodatabase>gene synonyms</aureodatabase> | *<aureodatabase>gene synonyms</aureodatabase> | ||
* <aureodatabase>product</aureodatabase> | *<aureodatabase>product</aureodatabase> | ||
</protect> | </protect> | ||
| Line 24: | Line 25: | ||
==General== | ==General== | ||
* <aureodatabase>gene type</aureodatabase> | *<aureodatabase>gene type</aureodatabase> | ||
* <aureodatabase>locus</aureodatabase> | *<aureodatabase>locus</aureodatabase> | ||
* <aureodatabase>gene symbol</aureodatabase> | *<aureodatabase>gene symbol</aureodatabase> | ||
* <aureodatabase>product</aureodatabase> | *<aureodatabase>product</aureodatabase> | ||
* <aureodatabase>gene replicon</aureodatabase> | *<aureodatabase>gene replicon</aureodatabase> | ||
* <aureodatabase>strand</aureodatabase> | *<aureodatabase>strand</aureodatabase> | ||
* <aureodatabase>gene coordinates</aureodatabase> | *<aureodatabase>gene coordinates</aureodatabase> | ||
* <aureodatabase>gene length</aureodatabase> | *<aureodatabase>gene length</aureodatabase> | ||
* <aureodatabase>essential</aureodatabase> | *<aureodatabase>essential</aureodatabase> | ||
*<aureodatabase>gene comment</aureodatabase> | |||
</protect> | </protect> | ||
| Line 38: | Line 40: | ||
==Accession numbers== | ==Accession numbers== | ||
* <aureodatabase>gene GI</aureodatabase> | *<aureodatabase>gene GI</aureodatabase> | ||
* <aureodatabase>gene RefSeq</aureodatabase> | *<aureodatabase>gene RefSeq</aureodatabase> | ||
*<aureodatabase>gene BioCyc</aureodatabase> | |||
*<aureodatabase>gene MicrobesOnline</aureodatabase> | |||
</protect> | </protect> | ||
<protect> | <protect> | ||
==Phenotype== | ==Phenotype== | ||
</protect> | </protect> | ||
Share your knowledge and add information here. [<span class="plainlinks">[//aureowiki.med.uni-greifswald.de/index.php?title={{PAGENAMEE}}&veaction=edit§ion=6 edit]</span>] | |||
<protect> | <protect> | ||
==DNA sequence== | ==DNA sequence== | ||
* <aureodatabase>gene sequence</aureodatabase> | *<aureodatabase>gene sequence</aureodatabase> | ||
</protect> | </protect> | ||
<protect> | <protect> | ||
<aureodatabase>RNA regulated operons</aureodatabase> | |||
</protect> | |||
<protect> | |||
=Protein= | =Protein= | ||
<aureodatabase>protein 3D view</aureodatabase> | <aureodatabase>protein 3D view</aureodatabase> | ||
==General== | ==General== | ||
* <aureodatabase>locus</aureodatabase> | *<aureodatabase>locus</aureodatabase> | ||
* <aureodatabase>protein symbol</aureodatabase> | *<aureodatabase>protein symbol</aureodatabase> | ||
* <aureodatabase>protein description</aureodatabase> | *<aureodatabase>protein description</aureodatabase> | ||
* <aureodatabase>protein length</aureodatabase> | *<aureodatabase>protein length</aureodatabase> | ||
* <aureodatabase>theoretical pI</aureodatabase> | *<aureodatabase>theoretical pI</aureodatabase> | ||
* <aureodatabase>theoretical MW</aureodatabase> | *<aureodatabase>theoretical MW</aureodatabase> | ||
* <aureodatabase>GRAVY</aureodatabase> | *<aureodatabase>GRAVY</aureodatabase> | ||
</protect> | </protect> | ||
| Line 71: | Line 78: | ||
==Function== | ==Function== | ||
* <aureodatabase>protein reaction</aureodatabase> | *<aureodatabase>protein reaction</aureodatabase> | ||
* <aureodatabase>protein TIGRFAM</aureodatabase> | *<aureodatabase>protein TIGRFAM</aureodatabase> | ||
* <aureodatabase>protein TheSeed</aureodatabase> | *<aureodatabase>protein TheSeed</aureodatabase> | ||
* <aureodatabase>protein PFAM</aureodatabase> | *<aureodatabase>protein PFAM</aureodatabase> | ||
</protect> | </protect> | ||
<protect> | <protect> | ||
==Structure, modifications & | ==Structure, modifications & cofactors== | ||
* <aureodatabase>protein domains</aureodatabase> | *<aureodatabase>protein domains</aureodatabase> | ||
* <aureodatabase>protein modifications</aureodatabase> | *<aureodatabase>protein modifications</aureodatabase> | ||
* <aureodatabase>protein cofactors</aureodatabase> | *<aureodatabase>protein cofactors</aureodatabase> | ||
* <aureodatabase>protein effectors</aureodatabase> | *<aureodatabase>protein effectors</aureodatabase> | ||
* <aureodatabase>protein | *<aureodatabase>protein regulated operons</aureodatabase> | ||
</protect> | </protect> | ||
| Line 90: | Line 97: | ||
==Localization== | ==Localization== | ||
* <aureodatabase>protein Psortb</aureodatabase> | *<aureodatabase>protein Psortb</aureodatabase> | ||
* <aureodatabase>protein LocateP</aureodatabase> | *<aureodatabase>protein DeepLocPro</aureodatabase> | ||
* <aureodatabase>protein SignalP</aureodatabase> | *<aureodatabase>protein LocateP</aureodatabase> | ||
* <aureodatabase>protein TMHMM</aureodatabase> | *<aureodatabase>protein SignalP</aureodatabase> | ||
*<aureodatabase>protein TMHMM</aureodatabase> | |||
</protect> | </protect> | ||
| Line 99: | Line 107: | ||
==Accession numbers== | ==Accession numbers== | ||
* <aureodatabase>protein GI</aureodatabase> | *<aureodatabase>protein GI</aureodatabase> | ||
* <aureodatabase>protein | *<aureodatabase>protein RefSeq</aureodatabase> | ||
* <aureodatabase>protein | *<aureodatabase>protein UniProt</aureodatabase> | ||
* <aureodatabase>protein | *<aureodatabase>protein STRING</aureodatabase> | ||
</protect> | </protect> | ||
| Line 108: | Line 116: | ||
==Protein sequence== | ==Protein sequence== | ||
* <aureodatabase>protein sequence</aureodatabase> | *<aureodatabase>protein sequence</aureodatabase> | ||
</protect> | </protect> | ||
<protect> | <protect> | ||
== | ==Experimental data== | ||
* <aureodatabase>protein validated peptides</aureodatabase> | *<aureodatabase>protein validated peptides</aureodatabase> | ||
*<aureodatabase>protein validated localization</aureodatabase> | |||
*<aureodatabase>protein validated quantitative data</aureodatabase> | |||
*<aureodatabase>protein partners</aureodatabase> | |||
</protect> | </protect> | ||
| Line 125: | Line 136: | ||
==Operon== | ==Operon== | ||
* <aureodatabase>operons</aureodatabase> | *<aureodatabase>operons</aureodatabase> | ||
</protect> | </protect> | ||
| Line 131: | Line 142: | ||
==Regulation== | ==Regulation== | ||
*<aureodatabase>regulators</aureodatabase> | |||
* <aureodatabase>regulators</aureodatabase> | |||
</protect> | </protect> | ||
| Line 138: | Line 148: | ||
==Transcription pattern== | ==Transcription pattern== | ||
* <aureodatabase>expression browser</aureodatabase> | *<aureodatabase>expression browser</aureodatabase> | ||
</protect> | </protect> | ||
| Line 144: | Line 154: | ||
==Protein synthesis (provided by Aureolib)== | ==Protein synthesis (provided by Aureolib)== | ||
* <aureodatabase>protein synthesis Aureolib</aureodatabase> | *<aureodatabase>protein synthesis Aureolib</aureodatabase> | ||
</protect> | </protect> | ||
<protect> | <protect> | ||
== | ==Protein stability== | ||
* <aureodatabase>protein half-life</aureodatabase> | *<aureodatabase>protein half-life</aureodatabase> | ||
</protect> | </protect> | ||
Latest revision as of 06:33, 11 March 2016
PangenomeCOLN315NCTC8325NewmanUSA300_FPR375704-0298108BA0217611819-97685071193ECT-R 2ED133ED98HO 5096 0412JH1JH9JKD6008JKD6159JSNZLGA251M013MRSA252MSHR1132MSSA476MW2Mu3Mu50RF122ST398T0131TCH60TW20USA300_TCH1516VC40
NCBI: 03-AUG-2016
⊟Summary[edit | edit source]
- organism: Staphylococcus aureus NCTC8325
- locus tag: SAOUHSC_02657
- pan locus tag?: SAUPAN005893000
- symbol: SAOUHSC_02657
- pan gene symbol?: —
- synonym:
- product: hypothetical protein
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
- type: CDS
- locus tag: SAOUHSC_02657
- symbol: SAOUHSC_02657
- product: hypothetical protein
- replicon: chromosome
- strand: +
- coordinates: 2441636..2441728
- length: 93
- essential: no DEG
⊟Accession numbers[edit | edit source]
- Gene ID: 3921219 NCBI
- RefSeq: YP_501119 NCBI
- BioCyc: G1I0R-2504 BioCyc
- MicrobesOnline: 1291090 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61GTGACAAAAAAGCGAAGATATGACACAACTGAATTTGGTTTAGCACATAGTATGACAGCT
AAAATAACTTTACATCAAGCGCTATACAAATAA60
93
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SAOUHSC_02657
- symbol: SAOUHSC_02657
- description: hypothetical protein
- length: 30
- theoretical pI: 10.5736
- theoretical MW: 3541.13
- GRAVY: -0.696667
⊟Function[edit | edit source]
- TIGRFAM:
- TheSEED:
- PFAM:
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors:
- effectors:
⊟Localization[edit | edit source]
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MTKKRRYDTTEFGLAHSMTAKITLHQALYK
⊟Experimental data[edit | edit source]
- experimentally validated:
- protein localization:
- quantitative data / protein copy number per cell:
- interaction partners:
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
- MicrobesOnline: no polycistronic organisation predicted
⊟Regulation[edit | edit source]
- regulator:
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: [1]
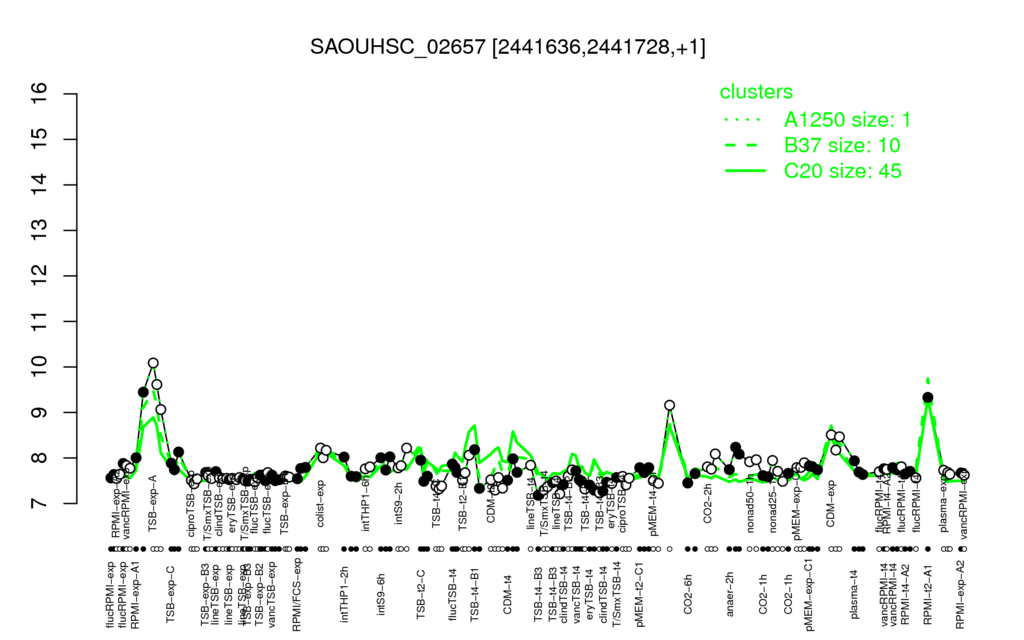 ⊟Multi-gene expression profiles
⊟Multi-gene expression profiles
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
- Aureolib: no data available
⊟Protein stability[edit | edit source]
- half-life: no data available
⊞Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊞Other Information[edit | edit source]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ Ulrike Mäder, Pierre Nicolas, Maren Depke, Jan Pané-Farré, Michel Debarbouille, Magdalena M van der Kooi-Pol, Cyprien Guérin, Sandra Dérozier, Aurelia Hiron, Hanne Jarmer, Aurélie Leduc, Stephan Michalik, Ewoud Reilman, Marc Schaffer, Frank Schmidt, Philippe Bessières, Philippe Noirot, Michael Hecker, Tarek Msadek, Uwe Völker, Jan Maarten van Dijl
Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions.
PLoS Genet: 2016, 12(4);e1005962
[PubMed:27035918] [WorldCat.org] [DOI] (I e)
