Jump to navigation
Jump to search
NCBI: 03-AUG-2016
⊟Summary[edit | edit source]
- organism: Staphylococcus aureus NCTC8325
- locus tag: SAOUHSC_00113
- pan locus tag?: SAUPAN000972000
- symbol: SAOUHSC_00113
- pan gene symbol?: adhE
- synonym:
- product: bifunctional acetaldehyde-CoA/alcohol dehydrogenase
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
- type: CDS
- locus tag: SAOUHSC_00113
- symbol: SAOUHSC_00113
- product: bifunctional acetaldehyde-CoA/alcohol dehydrogenase
- replicon: chromosome
- strand: +
- coordinates: 116538..119147
- length: 2610
- essential: no DEG other strains
⊟Accession numbers[edit | edit source]
- Gene ID: 3919822 NCBI
- RefSeq: YP_498713 NCBI
- BioCyc: G1I0R-104 BioCyc
- MicrobesOnline: 1288607 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421
481
541
601
661
721
781
841
901
961
1021
1081
1141
1201
1261
1321
1381
1441
1501
1561
1621
1681
1741
1801
1861
1921
1981
2041
2101
2161
2221
2281
2341
2401
2461
2521
2581ATGTTAACTATACCTGAAAAAGAAAATCGTGGATCGAAAGAACAAGAAGTGGCAATTATG
ATTGATGCTCTAGCTGACAAAGGGAAAAAAGCATTAGAAGCATTATCTAAAAAGTCACAA
GAAGAAATTGATCATATTGTTCATCAAATGAGCTTAGCAGCTGTTGATCAACATATGGTG
CTAGCAAAATTAGCACATGAAGAAACTGGAAGAGGTATATACGAAGATAAAGCGATTAAA
AATTTATACGCTTCTGAATATATATGGAATTCAATAAAAGACAATAAGACAGTAGGGATT
ATTGGTGAAGATAAAGAAAAAGGATTAACGTATGTAGCGGAACCAATTGGTGTTATTTGT
GGTGTTACGCCAACAACAAATCCTACGTCGACAACTATTTTTAAAGCGATGATTGCAATT
AAGACAGGAAATCCAATCATTTTTGCATTCCATCCAAGTGCACAAGAATCGTCGAAGCGT
GCAGCAGAAGTTGTATTAGAAGCGGCAATGAAGGCAGGTGCACCTAAAGATATTATTCAG
TGGATTGAAGTGCCTTCTATCGAAGCAACAAAACAATTAATGAATCACAAAGGTATTGCA
TTAGTTCTAGCAACAGGTGGTTCGGGCATGGTTAAGTCTGCATATTCAACTGGCAAACCG
GCATTAGGTGTGGGACCAGGTAACGTGCCGTCTTACATTGAAAAAACAGCACACATTAAA
CGTGCAGTAAATGATATCATTGGTTCAAAAACATTTGATAATGGTATGATTTGTGCTTCT
GAACAAGTTGTAGTCATTGATAAAGAAATTTATAAAGATGTTACTAATGAATTTAAAGCA
CATCAAGCATACTTTGTTAAAAAAGATGAATTACAACGCTTAGAAAATGCAATTATGAAT
GAACAAAAAACAGGTATTAAGCCTGATATTGTCGGTAAATCTGCAGTTGAAATAGCTGAA
TTAGCAGGTATACCTGTCCCCGAAAATACAAAACTTATCATAGCCGAAATTAGCGGTGTA
GGTTCAGACTATCCGTTATCTCGTGAAAAATTATCTCCAGTATTAGCCTTAGTAAAAGCC
CAATCTACAAAACAAGCATTTCAAATTTGTGAAGACACACTACATTTTGGTGGATTAGGA
CACACAGCCGTTATCCATACAGAAGATGAAACATTACAAAAAGATTTTGGACTAAGAATG
AAAGCTTGTCGTGTACTTGTAAATACACCATCAGCGGTTGGAGGTATTGGTGATATGTAT
AACGAATTGATTCCGTCTTTAACATTAGGTTGTGGTTCCTACGGTAGAAACTCAATTTCA
CATAATGTTAGTGCGACAGATTTATTAAACATTAAAACGATTGCTAAACGACGTAATAAT
ACTCAAATTTTCAAGGTGCCTGCTCAAATTTATTTTGAAGAAAATGCAATCATGAGTCTA
ACAACAATGGACAAGATTGAAAAAGTGATGATTGTCTGTGACCCTGGTATGGTAGAATTC
GGTTATACAAAAACAGTTGAGAATGTATTAAGACAAAAAACGGAACAGCCTCAAATAAAA
ATATTTAGCGAAGTCGAACCGAACCCATCAACTAATACAGTATATAAAGGTCTGGAAATG
ATGGTTGATTTCCAACCGGATACAATCATTGCACTTGGTGGTGGTTCAGCGATGGATGCT
GCAAAAGCAATGTGGATGTTCTTTGAACACCCTGAGACATCATTCTTCGGTGCTAAACAA
AAGTTCCTAGACATCGGTAAACGTACTTATAAAATAGGCATGCCTGAAAATGCGACGTTC
ATTTGTATCCCTACGACATCAGGTACAGGTTCAGAAGTAACACCATTTGCAGTTATCACA
GATAGTGAAACAAATGTAAAATATCCGTTGGCTGATTTTGCTTTAACACCTGACGTTGCA
ATTATTGACCCTCAATTTGTGATGAGTGTGCCAAAAAGCGTTACAGCAGATACAGGAATG
GATGTACTAACGCATGCAATGGAATCATATGTATCTGTAATGGCTTCAGACTACACAAGA
GGTTTGAGTCTACAAGCGATTAAATTGACGTTCGAATATTTAAAATCATCTGTTGAAAAG
GGTGATAAAGTTTCAAGAGAGAAAATGCATAACGCATCAACTTTGGCTGGTATGGCATTT
GCAAATGCATTCTTAGGCATTGCACACTCAATTGCGCATAAAATTGGTGGCGAATATGGT
ATTCCGCATGGTAGAGCGAATGCGATATTACTACCGCATATTATCCGTTATAATGCCAAA
GACCCGCAAAAACATGCATTATTCCCTAAATATGAGTTCTTCAGAGCAGATACAGATTAT
GCAGATATTGCCAAATTCTTAGGATTAAAAGGTAATACGACAGAAGCACTCGTAGAATCA
TTAGCTAAAGCTGTCTACGAATTAGGTCAATCAGTCGGAATTGAAATGAATTTGAAATCA
CAAGGTGTGTCTGAAGAAGAATTAAATGAGTCAATTGATAGAATGGCAGAGCTCGCATTT
GAAGATCAATGTACAACTGCTAATCCTAAAGAAGCACTAATCAGTGAAATCAAAGATATC
ATTCAAACATCATATGATTATAAGCAATAA60
120
180
240
300
360
420
480
540
600
660
720
780
840
900
960
1020
1080
1140
1200
1260
1320
1380
1440
1500
1560
1620
1680
1740
1800
1860
1920
1980
2040
2100
2160
2220
2280
2340
2400
2460
2520
2580
2610
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SAOUHSC_00113
- symbol: SAOUHSC_00113
- description: bifunctional acetaldehyde-CoA/alcohol dehydrogenase
- length: 869
- theoretical pI: 5.84639
- theoretical MW: 94944.4
- GRAVY: -0.149252
⊟Function[edit | edit source]
- TIGRFAM: acetaldehyde dehydrogenase (acetylating) (TIGR02518; EC 1.2.1.10; HMM-score: 514.9)and 13 moreEnergy metabolism Sugars lactaldehyde reductase (TIGR02638; EC 1.1.1.77; HMM-score: 280)phosphonate metabolism-associated iron-containing alcohol dehydrogenase (TIGR03405; HMM-score: 192.2)NDMA-dependent methanol dehydrogenase (TIGR04266; EC 1.1.99.37; HMM-score: 109.3)Amino acid biosynthesis Glutamate family glutamate-5-semialdehyde dehydrogenase (TIGR00407; EC 1.2.1.41; HMM-score: 55.2)Cellular processes Adaptations to atypical conditions betaine-aldehyde dehydrogenase (TIGR01804; EC 1.2.1.8; HMM-score: 55.2)Central intermediary metabolism Other succinate-semialdehyde dehydrogenase (TIGR01780; EC 1.2.1.-; HMM-score: 54.5)5-carboxymethyl-2-hydroxymuconate semialdehyde dehydrogenase (TIGR02299; EC 1.2.1.60; HMM-score: 39.6)Energy metabolism Amino acids and amines succinylglutamate-semialdehyde dehydrogenase (TIGR03240; EC 1.2.1.71; HMM-score: 37.5)Energy metabolism Amino acids and amines methylmalonate-semialdehyde dehydrogenase (acylating) (TIGR01722; EC 1.2.1.27; HMM-score: 33.8)putative phosphonoacetaldehyde dehydrogenase (TIGR03250; EC 1.2.1.-; HMM-score: 25.3)Energy metabolism Other 2-hydroxymuconic semialdehyde dehydrogenase (TIGR03216; EC 1.2.1.-; HMM-score: 24.6)Energy metabolism Amino acids and amines putative delta-1-pyrroline-5-carboxylate dehydrogenase (TIGR01237; EC 1.2.1.88; HMM-score: 24.1)Unknown function Enzymes of unknown specificity aldehyde dehydrogenase, Rv0768 family (TIGR04284; EC 1.2.1.-; HMM-score: 14.9)
- TheSEED :
- Acetaldehyde dehydrogenase (EC 1.2.1.10)
- Alcohol dehydrogenase (EC 1.1.1.1)
Carbohydrates Central carbohydrate metabolism Pyruvate metabolism II: acetyl-CoA, acetogenesis from pyruvate Acetaldehyde dehydrogenase (EC 1.2.1.10)and 5 more - PFAM: ADH_DHQS_C-like (CL0886) ADH_Fe_C; Fe-containing alcohol dehydrogenase family, C-terminal (PF25137; HMM-score: 215.7)and 3 moreDHQS (CL0224) Fe-ADH; Iron-containing alcohol dehydrogenase (PF00465; HMM-score: 155.5)ALDH-like (CL0099) Aldedh; Aldehyde dehydrogenase family (PF00171; HMM-score: 93.3)DHQS (CL0224) Fe-ADH_2; Iron-containing alcohol dehydrogenase (PF13685; HMM-score: 23.9)
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors:
- effectors:
⊟Localization[edit | edit source]
- PSORTb: Cytoplasmic
- Cytoplasmic Score: 9.97
- Cytoplasmic Membrane Score: 0
- Cellwall Score: 0.01
- Extracellular Score: 0.02
- Internal Helices: 0
- DeepLocPro: Cytoplasmic
- Cytoplasmic Score: 0.8098
- Cytoplasmic Membrane Score: 0.146
- Cell wall & surface Score: 0.0015
- Extracellular Score: 0.0427
- LocateP: Intracellular
- Prediction by SwissProt Classification: Cytoplasmic
- Pathway Prediction: No pathway
- Intracellular possibility: 1
- Signal peptide possibility: -1
- N-terminally Anchored Score: -1
- Predicted Cleavage Site: No CleavageSite
- SignalP: no predicted signal peptide
- SP(Sec/SPI): 0.002759
- TAT(Tat/SPI): 0.000416
- LIPO(Sec/SPII): 0.000451
- predicted transmembrane helices (TMHMM): 0
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MLTIPEKENRGSKEQEVAIMIDALADKGKKALEALSKKSQEEIDHIVHQMSLAAVDQHMVLAKLAHEETGRGIYEDKAIKNLYASEYIWNSIKDNKTVGIIGEDKEKGLTYVAEPIGVICGVTPTTNPTSTTIFKAMIAIKTGNPIIFAFHPSAQESSKRAAEVVLEAAMKAGAPKDIIQWIEVPSIEATKQLMNHKGIALVLATGGSGMVKSAYSTGKPALGVGPGNVPSYIEKTAHIKRAVNDIIGSKTFDNGMICASEQVVVIDKEIYKDVTNEFKAHQAYFVKKDELQRLENAIMNEQKTGIKPDIVGKSAVEIAELAGIPVPENTKLIIAEISGVGSDYPLSREKLSPVLALVKAQSTKQAFQICEDTLHFGGLGHTAVIHTEDETLQKDFGLRMKACRVLVNTPSAVGGIGDMYNELIPSLTLGCGSYGRNSISHNVSATDLLNIKTIAKRRNNTQIFKVPAQIYFEENAIMSLTTMDKIEKVMIVCDPGMVEFGYTKTVENVLRQKTEQPQIKIFSEVEPNPSTNTVYKGLEMMVDFQPDTIIALGGGSAMDAAKAMWMFFEHPETSFFGAKQKFLDIGKRTYKIGMPENATFICIPTTSGTGSEVTPFAVITDSETNVKYPLADFALTPDVAIIDPQFVMSVPKSVTADTGMDVLTHAMESYVSVMASDYTRGLSLQAIKLTFEYLKSSVEKGDKVSREKMHNASTLAGMAFANAFLGIAHSIAHKIGGEYGIPHGRANAILLPHIIRYNAKDPQKHALFPKYEFFRADTDYADIAKFLGLKGNTTEALVESLAKAVYELGQSVGIEMNLKSQGVSEEELNESIDRMAELAFEDQCTTANPKEALISEIKDIIQTSYDYKQ
⊟Experimental data[edit | edit source]
- experimentally validated: PeptideAtlas [1] [2]
- protein localization: data available for COL
- quantitative data / protein copy number per cell:
- interaction partners:
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
- MicrobesOnline: no polycistronic organisation predicted
⊟Regulation[edit | edit source]
- regulator: Rex* (repression) regulon
Rex* (TF) important in Energy metabolism; RegPrecise
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: [3]
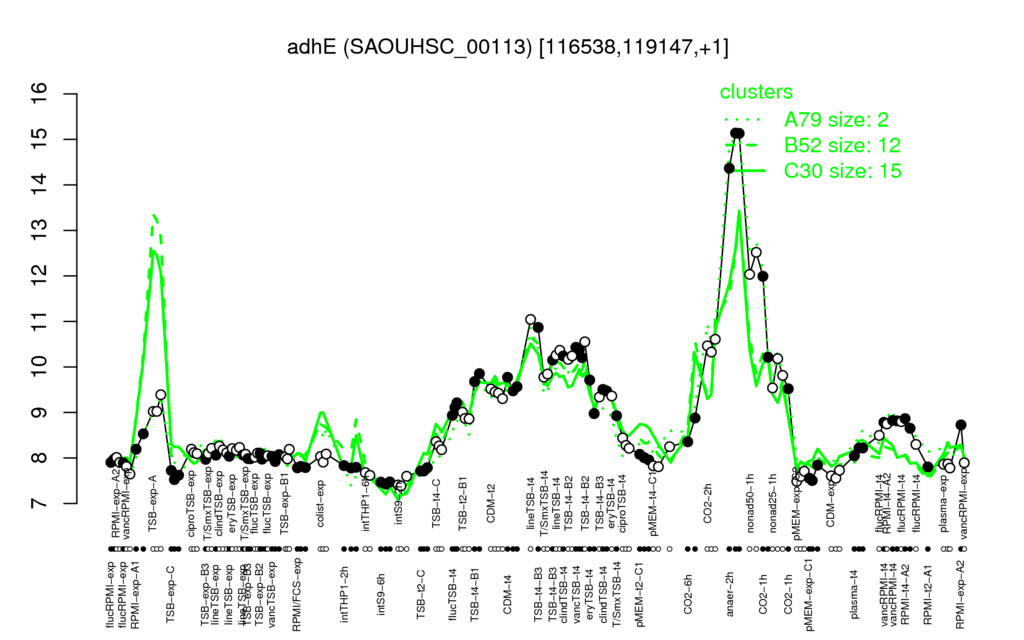 Multi-gene expression profiles
Multi-gene expression profiles
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
⊟Protein stability[edit | edit source]
- half-life: no data available
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You can add further information about the gene and protein here. [edit]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ Maren Depke, Stephan Michalik, Alexander Rabe, Kristin Surmann, Lars Brinkmann, Nico Jehmlich, Jörg Bernhardt, Michael Hecker, Bernd Wollscheid, Zhi Sun, Robert L Moritz, Uwe Völker, Frank Schmidt
A peptide resource for the analysis of Staphylococcus aureus in host-pathogen interaction studies.
Proteomics: 2015, 15(21);3648-61
[PubMed:26224020] [WorldCat.org] [DOI] (I p) - ↑ Stephan Michalik, Maren Depke, Annette Murr, Manuela Gesell Salazar, Ulrike Kusebauch, Zhi Sun, Tanja C Meyer, Kristin Surmann, Henrike Pförtner, Petra Hildebrandt, Stefan Weiss, Laura Marcela Palma Medina, Melanie Gutjahr, Elke Hammer, Dörte Becher, Thomas Pribyl, Sven Hammerschmidt, Eric W Deutsch, Samuel L Bader, Michael Hecker, Robert L Moritz, Ulrike Mäder, Uwe Völker, Frank Schmidt
A global Staphylococcus aureus proteome resource applied to the in vivo characterization of host-pathogen interactions.
Sci Rep: 2017, 7(1);9718
[PubMed:28887440] [WorldCat.org] [DOI] (I e) - ↑ Ulrike Mäder, Pierre Nicolas, Maren Depke, Jan Pané-Farré, Michel Debarbouille, Magdalena M van der Kooi-Pol, Cyprien Guérin, Sandra Dérozier, Aurelia Hiron, Hanne Jarmer, Aurélie Leduc, Stephan Michalik, Ewoud Reilman, Marc Schaffer, Frank Schmidt, Philippe Bessières, Philippe Noirot, Michael Hecker, Tarek Msadek, Uwe Völker, Jan Maarten van Dijl
Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions.
PLoS Genet: 2016, 12(4);e1005962
[PubMed:27035918] [WorldCat.org] [DOI] (I e)
