Jump to navigation
Jump to search
UniProt: 18-JAN-2017
⊟Summary[edit | edit source]
- organism: Staphylococcus aureus NCTC8325
- locus tag: SAOUHSC_00411.4
- pan locus tag?: SAUPAN002162000
- symbol: psmα1
- pan gene symbol?: psmα1
- synonym: psmA1
- product: Phenol-soluble modulin alpha 1 peptide
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
- type: CDS
- locus tag: SAOUHSC_00411.4
- symbol: psmα1
- product: Phenol-soluble modulin alpha 1 peptide
- replicon: chromosome
- strand: -
- coordinates: 412926..412991
- length: 66
- essential: unknown
⊟Accession numbers[edit | edit source]
- Gene ID:
- RefSeq:
- BioCyc: G1I0R-3544 BioCyc
- MicrobesOnline:
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61ATGGGTATCATCGCTGGCATCATTAAAGTTATCAAAAGCTTAATCGAACAATTCACTGGT
AAATAA60
66
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SAOUHSC_00411.4
- symbol: Psmα1
- description: Phenol-soluble modulin alpha 1 peptide
- length: 21
- theoretical pI: 10.5069
- theoretical MW: 2259.81
- GRAVY: 0.957143
⊟Function[edit | edit source]
- TIGRFAM:
- TheSEED:
- PFAM: no clan defined PSMalpha; Phenol-soluble modulin alpha peptide family (PF17063; HMM-score: 46)
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors:
- effectors:
⊟Localization[edit | edit source]
- PSORTb: unknown (no significant prediction)
- Cytoplasmic Score: 2.5
- Cytoplasmic Membrane Score: 2.5
- Cellwall Score: 2.5
- Extracellular Score: 2.5
- Internal Helices: 0
- DeepLocPro: Extracellular
- Cytoplasmic Score: 0
- Cytoplasmic Membrane Score: 0.0002
- Cell wall & surface Score: 0
- Extracellular Score: 0.9998
- LocateP:
- SignalP: no predicted signal peptide
- SP(Sec/SPI): 0.179334
- TAT(Tat/SPI): 0.019163
- LIPO(Sec/SPII): 0.091004
- predicted transmembrane helices (TMHMM): 0
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MGIIAGIIKVIKSLIEQFTGK
⊟Experimental data[edit | edit source]
- experimentally validated:
- protein localization:
- quantitative data / protein copy number per cell:
- interaction partners:
⊟Expression & Regulation[edit | edit source]
⊟Regulation[edit | edit source]
- regulator: AgrA* (activation) regulon
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: [3] The transcript corresponding to psmα1-4 mRNA was identified as new RNA feature (S130) in [3] because the psmα-genes are not covered by the RefSeq annotation.
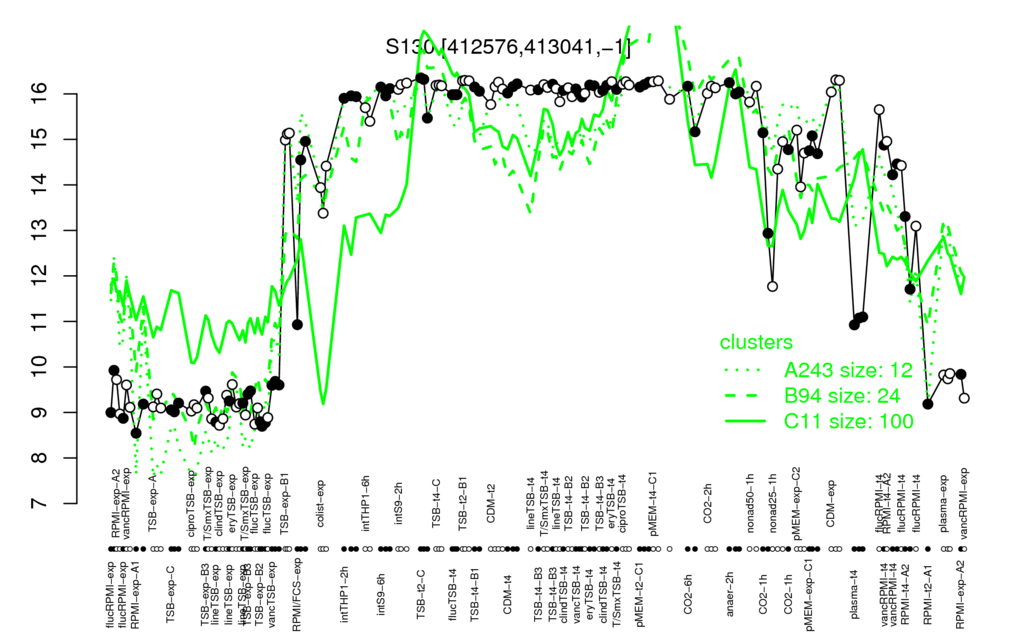 Multi-gene expression profiles
Multi-gene expression profiles
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
- Aureolib: no data available
⊟Protein stability[edit | edit source]
- half-life: no data available
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You can add further information about the gene and protein here. [edit]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ Shu Y Queck, Max Jameson-Lee, Amer E Villaruz, Thanh-Huy L Bach, Burhan A Khan, Daniel E Sturdevant, Stacey M Ricklefs, Min Li, Michael Otto
RNAIII-independent target gene control by the agr quorum-sensing system: insight into the evolution of virulence regulation in Staphylococcus aureus.
Mol Cell: 2008, 32(1);150-8
[PubMed:18851841] [WorldCat.org] [DOI] (I p) - ↑ Tobias Geiger, Patrice Francois, Manuel Liebeke, Martin Fraunholz, Christiane Goerke, Bernhard Krismer, Jacques Schrenzel, Michael Lalk, Christiane Wolz
The stringent response of Staphylococcus aureus and its impact on survival after phagocytosis through the induction of intracellular PSMs expression.
PLoS Pathog: 2012, 8(11);e1003016
[PubMed:23209405] [WorldCat.org] [DOI] (I p) - ↑ 3.0 3.1 Ulrike Mäder, Pierre Nicolas, Maren Depke, Jan Pané-Farré, Michel Debarbouille, Magdalena M van der Kooi-Pol, Cyprien Guérin, Sandra Dérozier, Aurelia Hiron, Hanne Jarmer, Aurélie Leduc, Stephan Michalik, Ewoud Reilman, Marc Schaffer, Frank Schmidt, Philippe Bessières, Philippe Noirot, Michael Hecker, Tarek Msadek, Uwe Völker, Jan Maarten van Dijl
Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions.
PLoS Genet: 2016, 12(4);e1005962
[PubMed:27035918] [WorldCat.org] [DOI] (I e)
⊟Relevant publications[edit | edit source]
Rong Wang, Kevin R Braughton, Dorothee Kretschmer, Thanh-Huy L Bach, Shu Y Queck, Min Li, Adam D Kennedy, David W Dorward, Seymour J Klebanoff, Andreas Peschel, Frank R DeLeo, Michael Otto
Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA.
Nat Med: 2007, 13(12);1510-4
[PubMed:17994102] [WorldCat.org] [DOI] (I p)Michael Otto
Staphylococcus aureus toxins.
Curr Opin Microbiol: 2014, 17;32-7
[PubMed:24581690] [WorldCat.org] [DOI] (I p)
