⊟Summary[edit | edit source]
- organism: Staphylococcus aureus NCTC8325
- locus tag: SAOUHSC_02798
- pan locus tag?: SAUPAN006108000
- symbol: SAOUHSC_02798
- pan gene symbol?: sasG
- synonym:
- product: hypothetical protein
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
- type: CDS
- locus tag: SAOUHSC_02798
- symbol: SAOUHSC_02798
- product: hypothetical protein
- replicon: chromosome
- strand: -
- coordinates: 2570009..2574892
- length: 4884
- essential: no DEG other strains
⊟Accession numbers[edit | edit source]
- Gene ID: 3921452 NCBI
- RefSeq: YP_501257 NCBI
- BioCyc: G1I0R-2636 BioCyc
- MicrobesOnline: 1291228 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421
481
541
601
661
721
781
841
901
961
1021
1081
1141
1201
1261
1321
1381
1441
1501
1561
1621
1681
1741
1801
1861
1921
1981
2041
2101
2161
2221
2281
2341
2401
2461
2521
2581
2641
2701
2761
2821
2881
2941
3001
3061
3121
3181
3241
3301
3361
3421
3481
3541
3601
3661
3721
3781
3841
3901
3961
4021
4081
4141
4201
4261
4321
4381
4441
4501
4561
4621
4681
4741
4801
4861ATGAGAGATAAGAAAGGACCGGTAAATAAAAGAGTAGATTTTCTATCAAATAAATTGAAT
AAATATTCAATAAGAAAATTTACAGTTGGAACAGCATCTATTTTAATTGGCTCACTAATG
TATTTGGGAACTCAACAAGAGGCAGAAGCAGCTGAAAACAATATTGAGAATCCAACTACA
TTAAAAGATAATGTCCAATCAAAAGAAGTGAAGATTGAAGAAGTAACAAACAAAGACACT
GCACCACAGGGTGTAGAAGCTAAATCTGAAGTAACTTCAAACAAAGACACAATCGAACAT
GAACCATCAGTAAAAGCTGAAGATATATCAAAAAAGGAGGATACACCAAAAGAAGTAGCT
GATGTTGCTGAAGTTCAGCCGAAATCGTCAGTCACTCATAACGCAGAGACACCTAAGGTT
AGAAAAGCTCGTTCTGTTGATGAAGGCTCTTTTGATATTACAAGAGATTCTAAAAATGTA
GTTGAATCTACCCCAATTACAATTCAAGGTAAAGAACATTTTGAAGGTTACGGAAGTGTT
GATATACAAAAAAAACCAACAGATTTAGGGGTATCAGAGGTAACCAGGTTTAATGTTGGT
AATGAAAGTAATGGTTTGATAGGAGCTTTACAATTAAAAAATAAAATAGATTTTAGTAAG
GATTTCAATTTTAAAGTTAGAGTGGCAAATAACCATCAATCAAATACCACAGGTGCTGAT
GGTTGGGGGTTCTTATTTAGTAAAGGAAATGCAGAAGAATATTTAACTAATGGTGGAATC
CTTGGGGATAAAGGTCTGGTAAATTCAGGCGGATTTAAAATTGATACTGGATACATTTAT
ACAAGTTCCATGGACAAAACTGAAAAGCAAGCTGGACAAGGTTATAGAGGATACGGAGCT
TTTGTGAAAAATGACAGTTCTGGTAATTCACAAATGGTTGGAGAAAATATTGATAAATCA
AAAACTAATTTTTTAAACTATGCGGACAATTCAACTAATACATCAGATGGAAAGTTTCAT
GGGCAACGTTTAAATGATGTCATCTTAACTTATGTTGCTTCAACTGGTAAAATGAGAGCA
GAATATGCTGGTAAAACTTGGGAGACTTCAATAACAGATTTAGGTTTATCTAAAAATCAG
GCATATAATTTCTTAATTACATCTAGTCAAAGATGGGGCCTTAATCAAGGGATAAATGCA
AATGGCTGGATGAGAACTGACTTGAAAGGTTCAGAGTTTACTTTTACACCAGAAGCGCCA
AAAACAATAACAGAATTAGAAAAAAAAGTTGAAGAGATTCCATTCAAGAAAGAACGTAAA
TTTAATCCGGATTTAGCACCAGGGACAGAAAAAGTAACAAGAGAAGGACAAAAAGGTGAG
AAGACAATAACGACACCAACACTAAAAAATCCATTAACTGGAGTAATTATTAGTAAAGGT
GAACCAAAAGAAGAGATTACAAAAGATCCGATTAATGAATTAACAGAATACGGACCTGAA
ACAATAGCGCCAGGTCATCGAGACGAATTTGATCCGAAGTTACCAACAGGAGAGAAAGAG
GAAGTTCCAGGTAAACCAGGAATTAAGAATCCAGAAACAGGAGACGTAGTTAGACCGCCG
GTCGATAGCGTAACAAAATATGGACCTGTAAAAGGAGACTCGATTGTAGAAAAAGAAGAG
ATTCCATTCGAGAAAGAACGTAAATTTAATCCTGATTTAGCACCAGGGACAGAAAAAGTA
ACAAGAGAAGGACAAAAAGGTGAGAAGACAATAACGACGCCAACACTAAAAAATCCATTA
ACTGGAGAAATTATTAGTAAAGGTGAATCGAAAGAAGAAATCACAAAAGATCCGATTAAT
GAATTAACAGAATACGGACCAGAAACGATAACACCAGGTCATCGAGACGAATTTGATCCG
AAGTTACCAACAGGAGAGAAAGAGGAAGTTCCAGGTAAACCAGGAATTAAGAATCCAGAA
ACAGGAGATGTAGTTAGACCACCGGTCGATAGCGTAACAAAATATGGACCTGTAAAAGGA
GACTCGATTGTAGAAAAAGAAGAGATTCCATTCGAGAAAGAACGTAAATTTAATCCTGAT
TTAGCACCAGGGACAGAAAAAGTAACAAGAGAAGGACAAAAAGGTGAGAAGACAATAACG
ACACCAACACTAAAAAATCCATTAACTGGAGTAATTATTAGTAAAGGTGAACCAAAAGAA
GAAATCACAAAAGATCCGATTAATGAATTAACAGAATACGGACCAGAAACGATAACACCA
GGTCATCGAGACGAATTTGATCCGAAGTTACCAACAGGAGAGAAAGAAGAAGTTCCAGGT
AAACCAGGAATTAAGAATCCAGAAACAGGAGACGTAGTTAGACCACCGGTCGATAGCGTA
ACAAAATATGGACCTGTAAAAGGAGACTCGATTGTAGAAAAAGAAGAGATTCCATTCAAG
AAAGAACGTAAATTTAATCCGGATTTAGCACCAGGGACAGAAAAAGTAACAAGAGAAGGA
CAAAAAGGTGAGAAGACAATAACGACGCCAACACTAAAAAATCCATTAACTGGAGAAATT
ATTAGTAAAGGTGAATCGAAAGAAGAAATCACAAAAGATCCGATTAATGAATTAACAGAA
TACGGACCAGAAACGATAACACCAGGTCATCGAGACGAATTTGATCCGAAGTTACCAACA
GGAGAGAAAGAGGAAGTTCCAGGTAAACCAGGAATTAAGAATCCAGAAACAGGAGATGTA
GTTAGACCACCGGTCGATAGCGTAACAAAATATGGACCTGTAAAAGGAGACTCGATTGTA
GAAAAAGAAGAGATTCCATTCGAGAAAGAACGTAAATTTAATCCTGATTTAGCACCAGGG
ACAGAAAAAGTAACAAGAGAAGGACAAAAAGGTGAGAAGACAATAACGACGCCAACACTA
AAAAATCCATTAACTGGAGAAATTATTAGTAAAGGTGAATCGAAAGAAGAAATCACAAAA
GATCCGATTAATGAATTAACAGAATACGGACCAGAAACGATAACACCAGGTCATCGAGAC
GAATTTGATCCGAAGTTACCAACAGGAGAGAAAGAGGAAGTTCCAGGTAAACCAGGAATT
AAGAATCCAGAAACAGGAGACGTAGTTAGACCACCGGTCGATAGCGTAACAAAATATGGA
CCTGTAAAAGGAGACTCGATTGTAGAAAAAGAAGAAATTCCATTCAAGAAAGAACGTAAA
TTTAATCCTGATTTAGCACCAGGGACAGAAAAAGTAACAAGAGAAGGACAAAAAGGTGAG
AAGACAATAACGACGCCAACACTAAAAAATCCATTAACTGGAGAAATTATTAGTAAAGGT
GAATCGAAAGAAGAAATCACAAAAGATCCGATTAATGAATTAACAGAATACGGACCAGAA
ACGATAACACCAGGTCATCGAGACGAATTTGATCCGAAGTTACCAACAGGAGAGAAAGAG
GAAGTTCCAGGTAAACCAGGAATTAAGAATCCAGAAACAGGAGATGTAGTTAGACCACCG
GTCGATAGCGTAACAAAATATGGACCTGTAAAAGGAGACTCGATTGTAGAAAAAGAAGAA
ATTCCATTCGAGAAAGAACGTAAATTTAATCCTGATTTAGCACCAGGGACAGAAAAAGTA
ACAAGAGAAGGACAAAAAGGTGAGAAGACAATAACGACGCCAACACTAAAAAATCCATTA
ACTGGAGAAATTATTAGTAAAGGTGAATCGAAAGAAGAAATCACAAAAGATCCGATTAAT
GAATTAACAGAATACGGACCAGAAACGATAACACCAGGTCATCGAGACGAATTTGATCCG
AAGTTACCAACAGGAGAGAAAGAGGAAGTTCCAGGTAAACCAGGAATTAAGAATCCAGAA
ACAGGAGATGTAGTTAGACCACCGGTCGATAGCGTAACAAAATATGGACCTGTAAAAGGA
GACTCGATTGTAGAAAAAGAAGAAATTCCATTCGAGAAAGAACGTAAATTTAATCCTGAT
TTAGCACCAGGGACAGAAAAAGTAACAAGAGAAGGACAAAAAGGTGAGAAGACAATAACG
ACGCCAACACTAAAAAATCCATTAACTGGAGAAATTATTAGTAAAGGTGAATCGAAAGAA
GAAATCACAAAAGATCCAGTTAATGAATTAACAGAATTCGGTGGCGAGAAAATACCGCAA
GGTCATAAAGATATCTTTGATCCAAACTTACCAACAGATCAAACGGAAAAAGTACCAGGT
AAACCAGGAATCAAGAATCCAGACACAGGAAAAGTGATCGAAGAGCCAGTGGATGATGTG
ATTAAACACGGACCAAAAACGGGTACACCAGAAACAAAAACAGTAGAGATACCGTTTGAA
ACAAAACGTGAGTTTAATCCAAAATTACAACCTGGTGAAGAGCGAGTGAAACAAGAAGGA
CAACCAGGAAGTAAGACAATCACAACACCAATCACAGTGAACCCATTAACAGGTGAAAAA
GTTGGCGAGGGTCAACCAACAGAAGAGATCACAAAACAACCAGTAGATAAGATTGTAGAG
TTCGGTGGAGAGAAACCAAAAGATCCAAAAGGACCTGAAAACCCAGAGAAGCCGAGCAGA
CCAACTCATCCAAGTGGCCCAGTAAATCCTAACAATCCAGGATTATCGAAAGACAGAGCA
AAACCAAATGGCCCAGTTCATTCAATGGATAAAAATGATAAAGTTAAAAAATCTAAAATT
GCTAAAGAATCAGTAGCTAATCAAGAGAAAAAACGAGCAGAATTACCAAAAACAGGTTTA
GAAAGCACGCAAAAAGGTTTGATCTTTAGTAGTATAATTGGAATTGCTGGATTAATGTTA
TTGGCTCGTAGAAGAAAGAATTAA60
120
180
240
300
360
420
480
540
600
660
720
780
840
900
960
1020
1080
1140
1200
1260
1320
1380
1440
1500
1560
1620
1680
1740
1800
1860
1920
1980
2040
2100
2160
2220
2280
2340
2400
2460
2520
2580
2640
2700
2760
2820
2880
2940
3000
3060
3120
3180
3240
3300
3360
3420
3480
3540
3600
3660
3720
3780
3840
3900
3960
4020
4080
4140
4200
4260
4320
4380
4440
4500
4560
4620
4680
4740
4800
4860
4884
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SAOUHSC_02798
- symbol: SAOUHSC_02798
- description: hypothetical protein
- length: 1627
- theoretical pI: 5.08263
- theoretical MW: 178526
- GRAVY: -0.986663
⊟Function[edit | edit source]
- TIGRFAM: calcium-translocating P-type ATPase, PMCA-type (TIGR01517; EC 3.6.3.8; HMM-score: 66)and 3 moregram-positive signal peptide, YSIRK family (TIGR01168; HMM-score: 46)Cell envelope Other LPXTG cell wall anchor domain (TIGR01167; HMM-score: 23.5)zinc metalloproteinase 18-residue repeat (TIGR04227; HMM-score: 8.3)
- TheSEED :
- Virulence-associated cell-wall-anchored protein SasG (LPXTG motif), binding to squamous nasal epithelial cells
- PFAM: G5 (CL0593) SasG_E; E domain (PF17041; HMM-score: 731.7)and 9 moreG5; G5 domain (PF07501; HMM-score: 382.8)Concanavalin (CL0004) Bact_lectin; Bacterial lectin (PF18483; HMM-score: 92.3)no clan defined YSIRK_signal; YSIRK type signal peptide (PF04650; HMM-score: 45.9)NTF2 (CL0051) DUF3828; Protein of unknown function (DUF3828) (PF12883; HMM-score: 40.1)DMP12-like (CL0859) DMP12; DNA-mimic protein (PF16779; HMM-score: 36.2)H-NS_histone_C (CL0786) MvaT_DBD; Bacterial xenogeneic silencer MvaT, C-terminal domain (PF22055; HMM-score: 27.7)no clan defined Gram_pos_anchor; LPXTG cell wall anchor motif (PF00746; HMM-score: 27)2C_adapt; 2-cysteine adaptor domain (PF08793; HMM-score: 22.2)Hybrid (CL0105) LAL_C2; L-amino acid ligase C-terminal domain 2 (PF18603; HMM-score: 12.7)
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors:
- effectors:
⊟Localization[edit | edit source]
- PSORTb: Cellwall
- Cytoplasmic Score: 0
- Cytoplasmic Membrane Score: 0
- Cellwall Score: 10
- Extracellular Score: 0
- Internal Helices: 0
- DeepLocPro: Cell wall & surface
- Cytoplasmic Score: 0.0003
- Cytoplasmic Membrane Score: 0.0036
- Cell wall & surface Score: 0.9153
- Extracellular Score: 0.0808
- LocateP: LPxTG Cell-wall anchored
- Prediction by SwissProt Classification: Cell Wall
- Pathway Prediction: Sec-(SPI)
- Intracellular possibility: 0.5
- Signal peptide possibility: -0.5
- N-terminally Anchored Score: 3
- Predicted Cleavage Site: found LPxTG motif :LPKTG
- SignalP: Signal peptide SP(Sec/SPI) length 50 aa
- SP(Sec/SPI): 0.831952
- TAT(Tat/SPI): 0.075519
- LIPO(Sec/SPII): 0.001918
- Cleavage Site: CS pos: 50-51. AEA-AE. Pr: 0.8873
- predicted transmembrane helices (TMHMM): 0
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MRDKKGPVNKRVDFLSNKLNKYSIRKFTVGTASILIGSLMYLGTQQEAEAAENNIENPTTLKDNVQSKEVKIEEVTNKDTAPQGVEAKSEVTSNKDTIEHEPSVKAEDISKKEDTPKEVADVAEVQPKSSVTHNAETPKVRKARSVDEGSFDITRDSKNVVESTPITIQGKEHFEGYGSVDIQKKPTDLGVSEVTRFNVGNESNGLIGALQLKNKIDFSKDFNFKVRVANNHQSNTTGADGWGFLFSKGNAEEYLTNGGILGDKGLVNSGGFKIDTGYIYTSSMDKTEKQAGQGYRGYGAFVKNDSSGNSQMVGENIDKSKTNFLNYADNSTNTSDGKFHGQRLNDVILTYVASTGKMRAEYAGKTWETSITDLGLSKNQAYNFLITSSQRWGLNQGINANGWMRTDLKGSEFTFTPEAPKTITELEKKVEEIPFKKERKFNPDLAPGTEKVTREGQKGEKTITTPTLKNPLTGVIISKGEPKEEITKDPINELTEYGPETIAPGHRDEFDPKLPTGEKEEVPGKPGIKNPETGDVVRPPVDSVTKYGPVKGDSIVEKEEIPFEKERKFNPDLAPGTEKVTREGQKGEKTITTPTLKNPLTGEIISKGESKEEITKDPINELTEYGPETITPGHRDEFDPKLPTGEKEEVPGKPGIKNPETGDVVRPPVDSVTKYGPVKGDSIVEKEEIPFEKERKFNPDLAPGTEKVTREGQKGEKTITTPTLKNPLTGVIISKGEPKEEITKDPINELTEYGPETITPGHRDEFDPKLPTGEKEEVPGKPGIKNPETGDVVRPPVDSVTKYGPVKGDSIVEKEEIPFKKERKFNPDLAPGTEKVTREGQKGEKTITTPTLKNPLTGEIISKGESKEEITKDPINELTEYGPETITPGHRDEFDPKLPTGEKEEVPGKPGIKNPETGDVVRPPVDSVTKYGPVKGDSIVEKEEIPFEKERKFNPDLAPGTEKVTREGQKGEKTITTPTLKNPLTGEIISKGESKEEITKDPINELTEYGPETITPGHRDEFDPKLPTGEKEEVPGKPGIKNPETGDVVRPPVDSVTKYGPVKGDSIVEKEEIPFKKERKFNPDLAPGTEKVTREGQKGEKTITTPTLKNPLTGEIISKGESKEEITKDPINELTEYGPETITPGHRDEFDPKLPTGEKEEVPGKPGIKNPETGDVVRPPVDSVTKYGPVKGDSIVEKEEIPFEKERKFNPDLAPGTEKVTREGQKGEKTITTPTLKNPLTGEIISKGESKEEITKDPINELTEYGPETITPGHRDEFDPKLPTGEKEEVPGKPGIKNPETGDVVRPPVDSVTKYGPVKGDSIVEKEEIPFEKERKFNPDLAPGTEKVTREGQKGEKTITTPTLKNPLTGEIISKGESKEEITKDPVNELTEFGGEKIPQGHKDIFDPNLPTDQTEKVPGKPGIKNPDTGKVIEEPVDDVIKHGPKTGTPETKTVEIPFETKREFNPKLQPGEERVKQEGQPGSKTITTPITVNPLTGEKVGEGQPTEEITKQPVDKIVEFGGEKPKDPKGPENPEKPSRPTHPSGPVNPNNPGLSKDRAKPNGPVHSMDKNDKVKKSKIAKESVANQEKKRAELPKTGLESTQKGLIFSSIIGIAGLMLLARRRKN
⊟Experimental data[edit | edit source]
- experimentally validated: PeptideAtlas [1] [2]
- protein localization:
- quantitative data / protein copy number per cell:
- interaction partners:
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
- predicted SigA promoter [3] : SAOUHSC_02798 < SAOUHSC_02799 < S1091
⊟Regulation[edit | edit source]
- regulator:
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: [3]
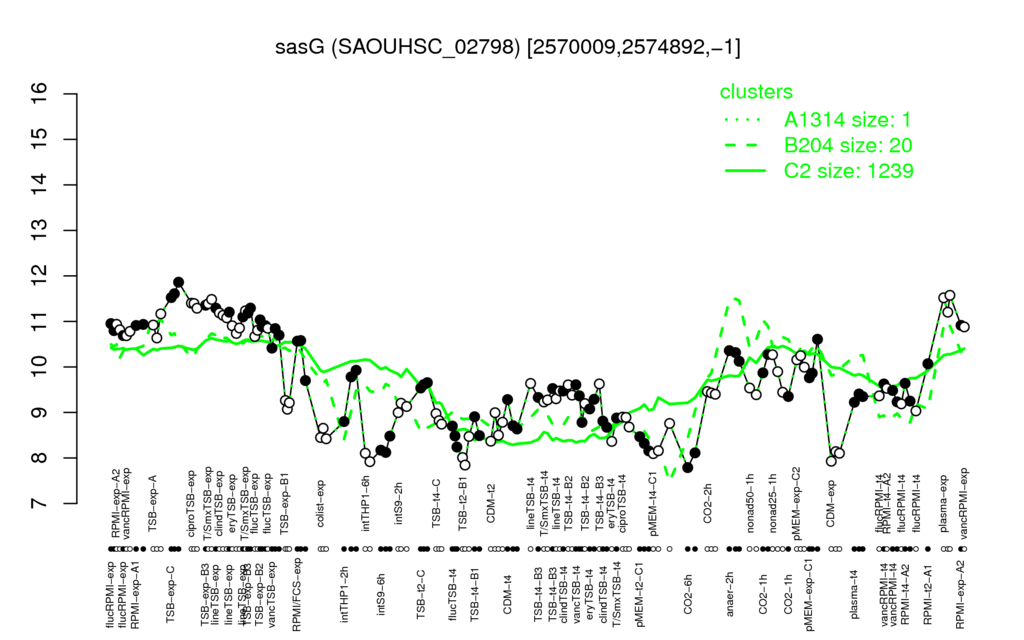 Multi-gene expression profiles
Multi-gene expression profiles
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
- Aureolib: no data available
⊟Protein stability[edit | edit source]
- half-life: no data available
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You can add further information about the gene and protein here. [edit]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ Maren Depke, Stephan Michalik, Alexander Rabe, Kristin Surmann, Lars Brinkmann, Nico Jehmlich, Jörg Bernhardt, Michael Hecker, Bernd Wollscheid, Zhi Sun, Robert L Moritz, Uwe Völker, Frank Schmidt
A peptide resource for the analysis of Staphylococcus aureus in host-pathogen interaction studies.
Proteomics: 2015, 15(21);3648-61
[PubMed:26224020] [WorldCat.org] [DOI] (I p) - ↑ Stephan Michalik, Maren Depke, Annette Murr, Manuela Gesell Salazar, Ulrike Kusebauch, Zhi Sun, Tanja C Meyer, Kristin Surmann, Henrike Pförtner, Petra Hildebrandt, Stefan Weiss, Laura Marcela Palma Medina, Melanie Gutjahr, Elke Hammer, Dörte Becher, Thomas Pribyl, Sven Hammerschmidt, Eric W Deutsch, Samuel L Bader, Michael Hecker, Robert L Moritz, Ulrike Mäder, Uwe Völker, Frank Schmidt
A global Staphylococcus aureus proteome resource applied to the in vivo characterization of host-pathogen interactions.
Sci Rep: 2017, 7(1);9718
[PubMed:28887440] [WorldCat.org] [DOI] (I e) - ↑ 3.0 3.1 Ulrike Mäder, Pierre Nicolas, Maren Depke, Jan Pané-Farré, Michel Debarbouille, Magdalena M van der Kooi-Pol, Cyprien Guérin, Sandra Dérozier, Aurelia Hiron, Hanne Jarmer, Aurélie Leduc, Stephan Michalik, Ewoud Reilman, Marc Schaffer, Frank Schmidt, Philippe Bessières, Philippe Noirot, Michael Hecker, Tarek Msadek, Uwe Völker, Jan Maarten van Dijl
Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions.
PLoS Genet: 2016, 12(4);e1005962
[PubMed:27035918] [WorldCat.org] [DOI] (I e)
⊟Relevant publications[edit | edit source]
Fiona M Roche, Ruth Massey, Sharon J Peacock, Nicholas P J Day, Livia Visai, Pietro Speziale, Alex Lam, Mark Pallen, Timothy J Foster
Characterization of novel LPXTG-containing proteins of Staphylococcus aureus identified from genome sequences.
Microbiology (Reading): 2003, 149(Pt 3);643-654
[PubMed:12634333] [WorldCat.org] [DOI] (P p)Fiona M Roche, Mary Meehan, Timothy J Foster
The Staphylococcus aureus surface protein SasG and its homologues promote bacterial adherence to human desquamated nasal epithelial cells.
Microbiology (Reading): 2003, 149(Pt 10);2759-2767
[PubMed:14523109] [WorldCat.org] [DOI] (P p)Rebecca M Corrigan, David Rigby, Pauline Handley, Timothy J Foster
The role of Staphylococcus aureus surface protein SasG in adherence and biofilm formation.
Microbiology (Reading): 2007, 153(Pt 8);2435-2446
[PubMed:17660408] [WorldCat.org] [DOI] (P p)Dominika T Gruszka, Justyna A Wojdyla, Richard J Bingham, Johan P Turkenburg, Iain W Manfield, Annette Steward, Andrew P Leech, Joan A Geoghegan, Timothy J Foster, Jane Clarke, Jennifer R Potts
Staphylococcal biofilm-forming protein has a contiguous rod-like structure.
Proc Natl Acad Sci U S A: 2012, 109(17);E1011-8
[PubMed:22493247] [WorldCat.org] [DOI] (I p)
