Jump to navigation
Jump to search
NCBI: 10-JUN-2013
⊟Summary[edit | edit source]
- organism: Staphylococcus aureus COL
- locus tag: SACOL1054 [new locus tag: SACOL_RS05375 ]
- pan locus tag?: SAUPAN003248000
- symbol: menB
- pan gene symbol?: menB
- synonym:
- product: naphthoate synthase
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
- type: CDS
- locus tag: SACOL1054 [new locus tag: SACOL_RS05375 ]
- symbol: menB
- product: naphthoate synthase
- replicon: chromosome
- strand: +
- coordinates: 1060326..1061147
- length: 822
- essential: unknown other strains
⊟Accession numbers[edit | edit source]
- Gene ID: 3236163 NCBI
- RefSeq: YP_185919 NCBI
- BioCyc: see SACOL_RS05375
- MicrobesOnline: 912523 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421
481
541
601
661
721
781ATGACTAACAGACAATGGGAAACACTTAGAGAATATGATGAAATCAAATATGAATTTTAC
GAAGGGATTGCTAAGGTAACAATAAATCGCCCTGAAGTACGCAATGCGTTTACACCTAAA
ACAGTTGCTGAAATGATTGACGCATTTTCACGTGCACGTGATGATCAAAACGTTTCAGTT
ATCGTATTAACTGGTGAAGGTGATTTAGCATTCTGTTCTGGTGGTGACCAGAAGAAACGT
GGACATGGTGGTTATGTAGGTGAAGACCAAATCCCTCGCTTAAATGTATTAGATTTACAG
CGTTTAATTCGTATTATTCCAAAACCGGTTATCGCGATGGTAAAAGGTTATGCTGTAGGT
GGCGGTAATGTACTAAATGTTGTTTGTGACTTAACGATTGCTGCTGATAATGCTATTTTT
GGACAAACTGGTCCTAAAGTAGGTTCATTTGATGCGGGTTATGGTTCAGGATATTTAGCA
CGTATCGTTGGACATAAGAAAGCACGTGAAATTTGGTACTTATGTCGTCAATACAATGCA
CAAGAAGCTTTAGATATGGGTCTAGTAAATACAGTGGTACCTTTAGAGAAAGTTGAAGAT
GAAACTGTGCAATGGTGTAAAGAGATTATGAAACACTCACCAACAGCGTTACGATTCCTT
AAAGCAGCTATGAATGCTGACACAGATGGTTTAGCTGGTTTACAACAAATGGCTGGGGAT
GCAACATTGCTTTATTACACAACTGATGAAGCGAAAGAAGGCCGTGATGCGTTTAAAGAA
AAACGTGATCCTGACTTCGATCAATTCCCTAAATTCCCATAA60
120
180
240
300
360
420
480
540
600
660
720
780
822
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SACOL1054 [new locus tag: SACOL_RS05375 ]
- symbol: MenB
- description: naphthoate synthase
- length: 273
- theoretical pI: 5.23939
- theoretical MW: 30425.4
- GRAVY: -0.358608
⊟Function[edit | edit source]
- reaction: EC 4.1.3.36? ExPASy1,4-dihydroxy-2-naphthoyl-CoA synthase 4-(2-carboxyphenyl)-4-oxobutanoyl-CoA = 1,4-dihydroxy-2-naphthoyl-CoA + H2O
- TIGRFAM: Biosynthesis of cofactors, prosthetic groups, and carriers Menaquinone and ubiquinone naphthoate synthase (TIGR01929; EC 4.1.3.36; HMM-score: 424.3)and 9 more2-ketocyclohexanecarboxyl-CoA hydrolase (TIGR03210; EC 3.7.-.-; HMM-score: 315.5)phenylacetate degradation probable enoyl-CoA hydratase PaaB (TIGR02280; EC 4.2.1.17; HMM-score: 118.4)6-oxocyclohex-1-ene-1-carbonyl-CoA hydrolase (TIGR03200; EC 3.-.-.-; HMM-score: 101.6)Fatty acid and phospholipid metabolism Degradation fatty oxidation complex, alpha subunit FadB (TIGR02437; EC 1.1.1.35,4.2.1.17,5.1.2.3,5.3.3.8; HMM-score: 56.8)cyclohexa-1,5-dienecarbonyl-CoA hydratase (TIGR03189; EC 4.2.1.100; HMM-score: 54.2)Fatty acid and phospholipid metabolism Degradation fatty oxidation complex, alpha subunit FadJ (TIGR02440; EC 1.1.1.35,4.2.1.17,5.1.2.3; HMM-score: 48.2)fatty acid oxidation complex, alpha subunit, mitochondrial (TIGR02441; EC 1.1.1.35,4.2.1.17; HMM-score: 36.6)benzoyl-CoA-dihydrodiol lyase (TIGR03222; EC 4.1.2.44; HMM-score: 21.1)Protein fate Degradation of proteins, peptides, and glycopeptides signal peptide peptidase SppA, 36K type (TIGR00706; EC 3.4.-.-; HMM-score: 13.5)
- TheSEED :
- Naphthoate synthase (EC 4.1.3.36)
Cofactors, Vitamins, Prosthetic Groups, Pigments Quinone cofactors Menaquinone and Phylloquinone Biosynthesis Naphthoate synthase (EC 4.1.3.36)and 1 more - PFAM: ClpP_crotonase (CL0127) ECH_1; Enoyl-CoA hydratase/isomerase (PF00378; HMM-score: 295.6)and 4 moreECH_2; Enoyl-CoA hydratase/isomerase (PF16113; HMM-score: 78.2)Peptidase_S49; Peptidase family S49 (PF01343; HMM-score: 13.6)no clan defined DUF1666; Protein of unknown function (DUF1666) (PF07891; HMM-score: 12.4)ClpP_crotonase (CL0127) SDH_protease; ClpP-like SDH-type serine proteinase (PF01972; HMM-score: 12.1)
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors: hydrogencarbonate
- effectors:
⊟Localization[edit | edit source]
- PSORTb: Cytoplasmic
- Cytoplasmic Score: 7.5
- Cytoplasmic Membrane Score: 1.15
- Cellwall Score: 0.62
- Extracellular Score: 0.73
- Internal Helices: 0
- DeepLocPro: Cytoplasmic
- Cytoplasmic Score: 0.9406
- Cytoplasmic Membrane Score: 0.0419
- Cell wall & surface Score: 0
- Extracellular Score: 0.0175
- LocateP: Intracellular
- Prediction by SwissProt Classification: Cytoplasmic
- Pathway Prediction: No pathway
- Intracellular possibility: 1
- Signal peptide possibility: -1
- N-terminally Anchored Score: 1
- Predicted Cleavage Site: No CleavageSite
- SignalP: no predicted signal peptide
- SP(Sec/SPI): 0.004575
- TAT(Tat/SPI): 0.000519
- LIPO(Sec/SPII): 0.000587
- predicted transmembrane helices (TMHMM): 0
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MTNRQWETLREYDEIKYEFYEGIAKVTINRPEVRNAFTPKTVAEMIDAFSRARDDQNVSVIVLTGEGDLAFCSGGDQKKRGHGGYVGEDQIPRLNVLDLQRLIRIIPKPVIAMVKGYAVGGGNVLNVVCDLTIAADNAIFGQTGPKVGSFDAGYGSGYLARIVGHKKAREIWYLCRQYNAQEALDMGLVNTVVPLEKVEDETVQWCKEIMKHSPTALRFLKAAMNADTDGLAGLQQMAGDATLLYYTTDEAKEGRDAFKEKRDPDFDQFPKFP
⊟Experimental data[edit | edit source]
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
- MicrobesOnline: SACOL1051 > menD > SACOL1053 > menB
⊟Regulation[edit | edit source]
- regulator:
⊟Transcription pattern[edit | edit source]
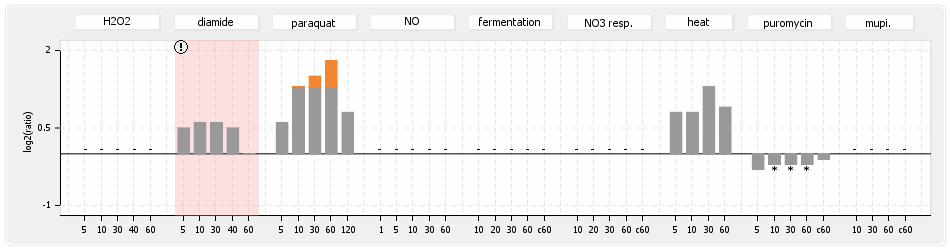
- S.aureus Expression Data Browser: data available for NCTC8325
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
⊟Protein stability[edit | edit source]
- half-life: no data available
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You can add further information about the gene and protein here. [edit]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ Dörte Becher, Kristina Hempel, Susanne Sievers, Daniela Zühlke, Jan Pané-Farré, Andreas Otto, Stephan Fuchs, Dirk Albrecht, Jörg Bernhardt, Susanne Engelmann, Uwe Völker, Jan Maarten van Dijl, Michael Hecker
A proteomic view of an important human pathogen--towards the quantification of the entire Staphylococcus aureus proteome.
PLoS One: 2009, 4(12);e8176
[PubMed:19997597] [WorldCat.org] [DOI] (I e) - ↑ Kristina Hempel, Jan Pané-Farré, Andreas Otto, Susanne Sievers, Michael Hecker, Dörte Becher
Quantitative cell surface proteome profiling for SigB-dependent protein expression in the human pathogen Staphylococcus aureus via biotinylation approach.
J Proteome Res: 2010, 9(3);1579-90
[PubMed:20108986] [WorldCat.org] [DOI] (I p) - ↑ Kristina Hempel, Florian-Alexander Herbst, Martin Moche, Michael Hecker, Dörte Becher
Quantitative proteomic view on secreted, cell surface-associated, and cytoplasmic proteins of the methicillin-resistant human pathogen Staphylococcus aureus under iron-limited conditions.
J Proteome Res: 2011, 10(4);1657-66
[PubMed:21323324] [WorldCat.org] [DOI] (I p) - ↑ Andreas Otto, Jan Maarten van Dijl, Michael Hecker, Dörte Becher
The Staphylococcus aureus proteome.
Int J Med Microbiol: 2014, 304(2);110-20
[PubMed:24439828] [WorldCat.org] [DOI] (I p) - ↑ Daniela Zühlke, Kirsten Dörries, Jörg Bernhardt, Sandra Maaß, Jan Muntel, Volkmar Liebscher, Jan Pané-Farré, Katharina Riedel, Michael Lalk, Uwe Völker, Susanne Engelmann, Dörte Becher, Stephan Fuchs, Michael Hecker
Costs of life - Dynamics of the protein inventory of Staphylococcus aureus during anaerobiosis.
Sci Rep: 2016, 6;28172
[PubMed:27344979] [WorldCat.org] [DOI] (I e)
⊟Relevant publications[edit | edit source]
Venkatasubramanian Ulaganathan, Mark F Agacan, Lori Buetow, Lindsay B Tulloch, William N Hunter
Structure of Staphylococcus aureus1,4-dihydroxy-2-naphthoyl-CoA synthase (MenB) in complex with acetoacetyl-CoA.
Acta Crystallogr Sect F Struct Biol Cryst Commun: 2007, 63(Pt 11);908-13
[PubMed:18007038] [WorldCat.org] [DOI] (I p)Ming Jiang, Minjiao Chen, Zu-Feng Guo, Zhihong Guo
A bicarbonate cofactor modulates 1,4-dihydroxy-2-naphthoyl-coenzyme a synthase in menaquinone biosynthesis of Escherichia coli.
J Biol Chem: 2010, 285(39);30159-69
[PubMed:20643650] [WorldCat.org] [DOI] (I p)