Jump to navigation
Jump to search
NCBI: 10-JUN-2013
⊟Summary[edit | edit source]
- organism: Staphylococcus aureus COL
- locus tag: SACOL1427 [new locus tag: SACOL_RS07270 ]
- pan locus tag?: SAUPAN003804000
- symbol: SACOL1427
- pan gene symbol?: —
- synonym:
- product: ABC transporter ATP-binding protein
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
- type: CDS
- locus tag: SACOL1427 [new locus tag: SACOL_RS07270 ]
- symbol: SACOL1427
- product: ABC transporter ATP-binding protein
- replicon: chromosome
- strand: +
- coordinates: 1438140..1439741
- length: 1602
- essential: unknown other strains
⊟Accession numbers[edit | edit source]
- Gene ID: 3236376 NCBI
- RefSeq: YP_186279 NCBI
- BioCyc: see SACOL_RS07270
- MicrobesOnline: 912885 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421
481
541
601
661
721
781
841
901
961
1021
1081
1141
1201
1261
1321
1381
1441
1501
1561ATGTTACAAGTAACTGATGTGAGTTTACGTTTTGGAGATCGTAAACTATTTGAAGATGTA
AATATTAAATTTACAGAAGGTAATTGTTATGGATTAATTGGTGCGAATGGTGCAGGTAAA
TCAACATTCTTAAAAATATTATCTGGTGAATTAGATTCTCAAACAGGACATGTTTCATTA
GGGAAAAATGAACGTCTAGCTGTTTTAAAACAGGACCACTATGCTTATGAAGATGAACGC
GTGCTTGATGTTGTAATTAAAGGTCACGAACGTCTTTATGAGGTTATGAAAGAAAAAGAT
GAAATCTATATGAAGCCAGATTTCAGTGATGAAGATGGTATCCGTGCTGCTGAACTTGAA
GGTGAATTTGCAGAAATGAATGGTTGGAATGCTGAAGCTGATGCTGCTAACCTTTTATCT
GGTTTAGGTATCGATCCAACTTTACACGATAAAAAAATGGCTGAATTAGAAAACAACCAA
AAAATTAAAGTATTATTAGCGCAAAGTTTATTCGGTGAACCAGACGTACTATTACTGGAT
GAGCCTACTAACGGTCTCGATATTCCAGCAATCAGTTGGTTAGAAGATTTCTTAATTAAC
TTTGATAATACTGTTATCGTAGTATCGCATGACCGTCATTTCTTAAATAATGTATGTACT
CATATCGCTGATTTAGACTTCGGTAAAATTAAAGTTTATGTTGGTAACTATGATTTTTGG
TATCAATCTAGTCAGTTAGCTCAAAAGATGGCTCAAGAACAAAACAAGAAAAAAGAAGAA
AAAATGAAAGAGTTACAGGACTTTATTGCTCGTTTCTCAGCTAACGCTTCTAAATCTAAA
CAAGCAACAAGTCGTAAAAAACAACTCGAGAAAATTGAATTAGATGATATTCAACCATCA
TCAAGAAGATATCCTTTCGTTAAATTCACACCTGAGCGCGAAATCGGTAATGACTTACTA
ATCGTTCAAAATCTATCTAAAACGATTGACGGTGAAAAAGTATTAGATAATATTTCATTC
ACAATGAATCCAAATGATAAAGCAATTTTAATTGGGGATAGTGAAATTGCGAAAACCACA
TTGCTTAAAATATTAGCCGGTGAAATGGAACCAGACGAAGGTTCATATAAATGGGGTGTA
ACAACGTCATTAAGTTACTTCCCTAAAGATAACTCAGAATTCTTTGAGGGCGTTAATATG
AACCTTGTAGATTGGTTAAGACAATACGCACCAGAAGATGAGCAAACTGAAACATTTTTA
CGCGGCTTCTTAGGCCGTATGCTATTTAGTGGAGAAGAAGTTAAGAAAAAAGCTAGTGTA
CTTTCAGGTGGAGAAAAAGTACGTTGTATGCTAAGTAAAATGATGTTATCAAGTGCAAAC
GTTCTTTTACTTGATGAACCAACAAACCACTTAGACTTAGAAAGTATTACTGCTGTTAAT
GATGGACTTAAATCATTCAAAGGTTCTATTATCTTTACTTCATATGACTTTGAATTTATC
AATACAATCGCAAACCGTGTTATCGATTTAAATAAACAAGGTGGCGTTTCAAAAGAAATT
CCATATGAAGAATATTTACAAGAAATCGGCGTTTTAAAATAA60
120
180
240
300
360
420
480
540
600
660
720
780
840
900
960
1020
1080
1140
1200
1260
1320
1380
1440
1500
1560
1602
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SACOL1427 [new locus tag: SACOL_RS07270 ]
- symbol: SACOL1427
- description: ABC transporter ATP-binding protein
- length: 533
- theoretical pI: 4.5829
- theoretical MW: 60255.9
- GRAVY: -0.383302
⊟Function[edit | edit source]
- TIGRFAM: ATP-binding cassette protein, ChvD family (TIGR03719; HMM-score: 311.2)and 71 moreproposed F420-0 ABC transporter, ATP-binding protein (TIGR03873; HMM-score: 138.2)thiol reductant ABC exporter, CydD subunit (TIGR02857; HMM-score: 137.9)Transport and binding proteins Unknown substrate energy-coupling factor transporter ATPase (TIGR04521; EC 3.6.3.-; HMM-score: 131.5)Transport and binding proteins Amino acids, peptides and amines urea ABC transporter, ATP-binding protein UrtD (TIGR03411; HMM-score: 128.8)gliding motility-associated ABC transporter ATP-binding subunit GldA (TIGR03522; HMM-score: 128.1)lantibiotic protection ABC transporter, ATP-binding subunit (TIGR03740; HMM-score: 122.8)Cellular processes Toxin production and resistance putative bacteriocin export ABC transporter, lactococcin 972 group (TIGR03608; HMM-score: 120.6)Transport and binding proteins Unknown substrate putative bacteriocin export ABC transporter, lactococcin 972 group (TIGR03608; HMM-score: 120.6)thiol reductant ABC exporter, CydC subunit (TIGR02868; HMM-score: 119.1)Transport and binding proteins Carbohydrates, organic alcohols, and acids ABC transporter, ATP-binding subunit, PQQ-dependent alcohol dehydrogenase system (TIGR03864; HMM-score: 118.9)Transport and binding proteins Other daunorubicin resistance ABC transporter, ATP-binding protein (TIGR01188; HMM-score: 118.7)Cellular processes Cell division cell division ATP-binding protein FtsE (TIGR02673; HMM-score: 115.6)Transport and binding proteins Anions phosphonate ABC transporter, ATP-binding protein (TIGR02315; EC 3.6.3.28; HMM-score: 115.4)Energy metabolism Methanogenesis methyl coenzyme M reductase system, component A2 (TIGR03269; HMM-score: 114.8)Protein fate Protein and peptide secretion and trafficking heme ABC exporter, ATP-binding protein CcmA (TIGR01189; EC 3.6.3.41; HMM-score: 113.4)Transport and binding proteins Other heme ABC exporter, ATP-binding protein CcmA (TIGR01189; EC 3.6.3.41; HMM-score: 113.4)Transport and binding proteins Cations and iron carrying compounds cobalt ABC transporter, ATP-binding protein (TIGR01166; HMM-score: 111.3)Protein fate Protein and peptide secretion and trafficking lipoprotein releasing system, ATP-binding protein (TIGR02211; EC 3.6.3.-; HMM-score: 106.9)Transport and binding proteins Amino acids, peptides and amines urea ABC transporter, ATP-binding protein UrtE (TIGR03410; HMM-score: 106.8)Transport and binding proteins Anions sulfate ABC transporter, ATP-binding protein (TIGR00968; EC 3.6.3.25; HMM-score: 105.9)Cellular processes Pathogenesis type I secretion system ATPase (TIGR03375; HMM-score: 104.9)Protein fate Protein and peptide secretion and trafficking type I secretion system ATPase (TIGR03375; HMM-score: 104.9)Transport and binding proteins Anions phosphate ABC transporter, ATP-binding protein (TIGR00972; EC 3.6.3.27; HMM-score: 99.8)ABC exporter ATP-binding subunit, DevA family (TIGR02982; HMM-score: 98)Transport and binding proteins Amino acids, peptides and amines putative 2-aminoethylphosphonate ABC transporter, ATP-binding protein (TIGR03265; HMM-score: 94)Transport and binding proteins Anions molybdate ABC transporter, ATP-binding protein (TIGR02142; EC 3.6.3.29; HMM-score: 93.9)Transport and binding proteins Carbohydrates, organic alcohols, and acids D-xylose ABC transporter, ATP-binding protein (TIGR02633; EC 3.6.3.17; HMM-score: 90.5)Transport and binding proteins Cations and iron carrying compounds nickel import ATP-binding protein NikE (TIGR02769; EC 3.6.3.24; HMM-score: 89.4)Biosynthesis of cofactors, prosthetic groups, and carriers Other FeS assembly ATPase SufC (TIGR01978; HMM-score: 88.6)Cell envelope Biosynthesis and degradation of surface polysaccharides and lipopolysaccharides LPS export ABC transporter ATP-binding protein (TIGR04406; HMM-score: 88)Transport and binding proteins Other LPS export ABC transporter ATP-binding protein (TIGR04406; HMM-score: 88)Protein fate Protein and peptide secretion and trafficking ABC-type bacteriocin transporter (TIGR01193; HMM-score: 87.5)Protein fate Protein modification and repair ABC-type bacteriocin transporter (TIGR01193; HMM-score: 87.5)Transport and binding proteins Other ABC-type bacteriocin transporter (TIGR01193; HMM-score: 87.5)Cellular processes Biosynthesis of natural products NHLM bacteriocin system ABC transporter, ATP-binding protein (TIGR03797; HMM-score: 86.4)Transport and binding proteins Amino acids, peptides and amines NHLM bacteriocin system ABC transporter, ATP-binding protein (TIGR03797; HMM-score: 86.4)Transport and binding proteins Unknown substrate energy-coupling factor transporter ATPase (TIGR04520; EC 3.6.3.-; HMM-score: 84.5)Cellular processes Other nodulation ABC transporter NodI (TIGR01288; HMM-score: 83.9)Transport and binding proteins Other nodulation ABC transporter NodI (TIGR01288; HMM-score: 83.9)Protein fate Protein and peptide secretion and trafficking type I secretion system ATPase (TIGR01842; HMM-score: 82.8)ectoine/hydroxyectoine ABC transporter, ATP-binding protein EhuA (TIGR03005; HMM-score: 82.5)phosphonate C-P lyase system protein PhnL (TIGR02324; HMM-score: 81.4)ABC transporter, permease/ATP-binding protein (TIGR02204; HMM-score: 79.9)Transport and binding proteins Unknown substrate anchored repeat-type ABC transporter, ATP-binding subunit (TIGR03771; HMM-score: 78.3)2-aminoethylphosphonate ABC transport system, ATP-binding component PhnT (TIGR03258; HMM-score: 73.4)Transport and binding proteins Other thiamine ABC transporter, ATP-binding protein (TIGR01277; EC 3.6.3.-; HMM-score: 72.5)Protein fate Protein and peptide secretion and trafficking type I secretion system ATPase (TIGR01846; HMM-score: 67.9)Cellular processes Biosynthesis of natural products NHLM bacteriocin system ABC transporter, peptidase/ATP-binding protein (TIGR03796; HMM-score: 66.6)Transport and binding proteins Amino acids, peptides and amines NHLM bacteriocin system ABC transporter, peptidase/ATP-binding protein (TIGR03796; HMM-score: 66.6)D-methionine ABC transporter, ATP-binding protein (TIGR02314; EC 3.6.3.-; HMM-score: 64.7)Transport and binding proteins Anions nitrate ABC transporter, ATP-binding proteins C and D (TIGR01184; HMM-score: 64.2)Transport and binding proteins Other nitrate ABC transporter, ATP-binding proteins C and D (TIGR01184; HMM-score: 64.2)Transport and binding proteins Amino acids, peptides and amines polyamine ABC transporter, ATP-binding protein (TIGR01187; EC 3.6.3.31; HMM-score: 63.5)Cell envelope Biosynthesis and degradation of surface polysaccharides and lipopolysaccharides lipid A export permease/ATP-binding protein MsbA (TIGR02203; HMM-score: 63.3)Transport and binding proteins Other lipid A export permease/ATP-binding protein MsbA (TIGR02203; HMM-score: 63.3)Transport and binding proteins Cations and iron carrying compounds nickel import ATP-binding protein NikD (TIGR02770; HMM-score: 63)Transport and binding proteins Amino acids, peptides and amines glycine betaine/L-proline transport ATP binding subunit (TIGR01186; HMM-score: 58.9)Transport and binding proteins Other antigen peptide transporter 2 (TIGR00958; HMM-score: 57.6)Central intermediary metabolism Phosphorus compounds phosphonate C-P lyase system protein PhnK (TIGR02323; HMM-score: 57.2)Transport and binding proteins Amino acids, peptides and amines choline ABC transporter, ATP-binding protein (TIGR03415; HMM-score: 55.1)Transport and binding proteins Anions cystic fibrosis transmembrane conductor regulator (CFTR) (TIGR01271; EC 3.6.3.49; HMM-score: 49.1)Transport and binding proteins Other pigment precursor permease (TIGR00955; HMM-score: 47.9)Transport and binding proteins Other rim ABC transporter (TIGR01257; HMM-score: 44.1)Transport and binding proteins Other pleiotropic drug resistance family protein (TIGR00956; HMM-score: 41)Transport and binding proteins Carbohydrates, organic alcohols, and acids glucan exporter ATP-binding protein (TIGR01192; EC 3.6.3.42; HMM-score: 38.8)Transport and binding proteins Other multi drug resistance-associated protein (MRP) (TIGR00957; HMM-score: 38)Transport and binding proteins Carbohydrates, organic alcohols, and acids peroxysomal long chain fatty acyl transporter (TIGR00954; HMM-score: 25.9)Transport and binding proteins Amino acids, peptides and amines cyclic peptide transporter (TIGR01194; HMM-score: 23.5)Transport and binding proteins Other cyclic peptide transporter (TIGR01194; HMM-score: 23.5)DNA metabolism DNA replication, recombination, and repair excinuclease ABC subunit A (TIGR00630; EC 3.1.25.-; HMM-score: 21.9)TIGR04168 family protein (TIGR04168; HMM-score: 12.4)
- TheSEED :
- ABC transporter ATP-binding protein uup
- PFAM: P-loop_NTPase (CL0023) ABC_tran; ABC transporter (PF00005; HMM-score: 140)and 18 moreABC_tran_Xtn; ABC transporter (PF12848; HMM-score: 76.8)AAA_21; AAA domain, putative AbiEii toxin, Type IV TA system (PF13304; HMM-score: 47.1)SMC_N; RecF/RecN/SMC N terminal domain (PF02463; HMM-score: 27.4)RsgA_GTPase; RsgA GTPase (PF03193; HMM-score: 24.4)AAA_29; P-loop containing region of AAA domain (PF13555; HMM-score: 22.9)AAA_23; AAA domain (PF13476; HMM-score: 19.2)MMR_HSR1; 50S ribosome-binding GTPase (PF01926; HMM-score: 14.9)AAA; ATPase family associated with various cellular activities (AAA) (PF00004; HMM-score: 14.6)AAA_22; AAA domain (PF13401; HMM-score: 14.3)Rad17; Rad17 P-loop domain (PF03215; HMM-score: 13.8)RNA_helicase; RNA helicase (PF00910; HMM-score: 13.7)AAA_18; AAA domain (PF13238; HMM-score: 13.7)VapE-like_dom; Virulence-associated protein E-like domain (PF05272; HMM-score: 13.4)HTH (CL0123) HTH_RNase_II; Double HTH domain (PF23161; HMM-score: 12.3)P-loop_NTPase (CL0023) RuvB_N; Holliday junction DNA helicase RuvB P-loop domain (PF05496; HMM-score: 12.2)AAA_24; AAA domain (PF13479; HMM-score: 11.3)DUF87; Helicase HerA, central domain (PF01935; HMM-score: 11.2)AAA_15; AAA ATPase domain (PF13175; HMM-score: 10.8)
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors:
- effectors:
⊟Localization[edit | edit source]
- PSORTb: Cytoplasmic
- Cytoplasmic Score: 9.97
- Cytoplasmic Membrane Score: 0
- Cellwall Score: 0.01
- Extracellular Score: 0.02
- Internal Helices: 0
- DeepLocPro: Cytoplasmic
- Cytoplasmic Score: 0.92
- Cytoplasmic Membrane Score: 0.0182
- Cell wall & surface Score: 0.0006
- Extracellular Score: 0.0612
- LocateP: Intracellular
- Prediction by SwissProt Classification: Cytoplasmic
- Pathway Prediction: No pathway
- Intracellular possibility: 1
- Signal peptide possibility: -1
- N-terminally Anchored Score: 1
- Predicted Cleavage Site: No CleavageSite
- SignalP: no predicted signal peptide
- SP(Sec/SPI): 0.059329
- TAT(Tat/SPI): 0.001081
- LIPO(Sec/SPII): 0.000694
- predicted transmembrane helices (TMHMM): 0
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MLQVTDVSLRFGDRKLFEDVNIKFTEGNCYGLIGANGAGKSTFLKILSGELDSQTGHVSLGKNERLAVLKQDHYAYEDERVLDVVIKGHERLYEVMKEKDEIYMKPDFSDEDGIRAAELEGEFAEMNGWNAEADAANLLSGLGIDPTLHDKKMAELENNQKIKVLLAQSLFGEPDVLLLDEPTNGLDIPAISWLEDFLINFDNTVIVVSHDRHFLNNVCTHIADLDFGKIKVYVGNYDFWYQSSQLAQKMAQEQNKKKEEKMKELQDFIARFSANASKSKQATSRKKQLEKIELDDIQPSSRRYPFVKFTPEREIGNDLLIVQNLSKTIDGEKVLDNISFTMNPNDKAILIGDSEIAKTTLLKILAGEMEPDEGSYKWGVTTSLSYFPKDNSEFFEGVNMNLVDWLRQYAPEDEQTETFLRGFLGRMLFSGEEVKKKASVLSGGEKVRCMLSKMMLSSANVLLLDEPTNHLDLESITAVNDGLKSFKGSIIFTSYDFEFINTIANRVIDLNKQGGVSKEIPYEEYLQEIGVLK
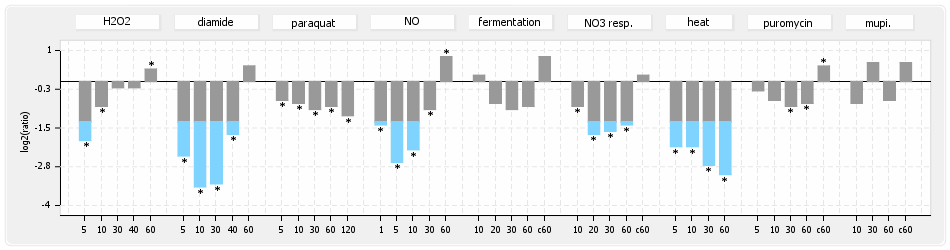
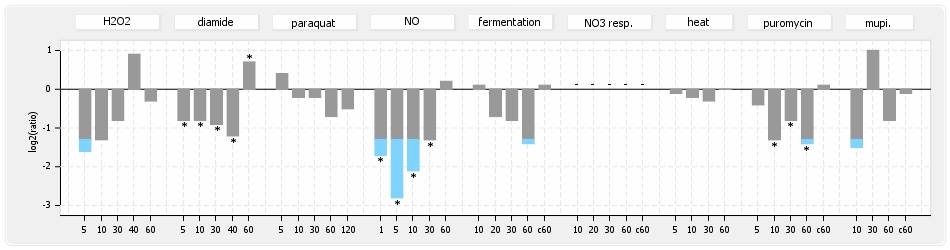
⊟Experimental data[edit | edit source]
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
- MicrobesOnline: no polycistronic organisation predicted
⊟Regulation[edit | edit source]
- regulator:
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: data available for NCTC8325
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
⊟Protein stability[edit | edit source]
- half-life: 59.21 h [6]
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You can add further information about the gene and protein here. [edit]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ Dörte Becher, Kristina Hempel, Susanne Sievers, Daniela Zühlke, Jan Pané-Farré, Andreas Otto, Stephan Fuchs, Dirk Albrecht, Jörg Bernhardt, Susanne Engelmann, Uwe Völker, Jan Maarten van Dijl, Michael Hecker
A proteomic view of an important human pathogen--towards the quantification of the entire Staphylococcus aureus proteome.
PLoS One: 2009, 4(12);e8176
[PubMed:19997597] [WorldCat.org] [DOI] (I e) - ↑ Kristina Hempel, Jan Pané-Farré, Andreas Otto, Susanne Sievers, Michael Hecker, Dörte Becher
Quantitative cell surface proteome profiling for SigB-dependent protein expression in the human pathogen Staphylococcus aureus via biotinylation approach.
J Proteome Res: 2010, 9(3);1579-90
[PubMed:20108986] [WorldCat.org] [DOI] (I p) - ↑ Kristina Hempel, Florian-Alexander Herbst, Martin Moche, Michael Hecker, Dörte Becher
Quantitative proteomic view on secreted, cell surface-associated, and cytoplasmic proteins of the methicillin-resistant human pathogen Staphylococcus aureus under iron-limited conditions.
J Proteome Res: 2011, 10(4);1657-66
[PubMed:21323324] [WorldCat.org] [DOI] (I p) - ↑ Andreas Otto, Jan Maarten van Dijl, Michael Hecker, Dörte Becher
The Staphylococcus aureus proteome.
Int J Med Microbiol: 2014, 304(2);110-20
[PubMed:24439828] [WorldCat.org] [DOI] (I p) - ↑ Daniela Zühlke, Kirsten Dörries, Jörg Bernhardt, Sandra Maaß, Jan Muntel, Volkmar Liebscher, Jan Pané-Farré, Katharina Riedel, Michael Lalk, Uwe Völker, Susanne Engelmann, Dörte Becher, Stephan Fuchs, Michael Hecker
Costs of life - Dynamics of the protein inventory of Staphylococcus aureus during anaerobiosis.
Sci Rep: 2016, 6;28172
[PubMed:27344979] [WorldCat.org] [DOI] (I e) - ↑ Stephan Michalik, Jörg Bernhardt, Andreas Otto, Martin Moche, Dörte Becher, Hanna Meyer, Michael Lalk, Claudia Schurmann, Rabea Schlüter, Holger Kock, Ulf Gerth, Michael Hecker
Life and death of proteins: a case study of glucose-starved Staphylococcus aureus.
Mol Cell Proteomics: 2012, 11(9);558-70
[PubMed:22556279] [WorldCat.org] [DOI] (I p)