⊟Summary[edit | edit source]
- organism: Staphylococcus aureus NCTC8325
- locus tag: SAOUHSC_00069
- pan locus tag?: SAUPAN000909000
- symbol: SAOUHSC_00069
- pan gene symbol?: spa
- synonym:
- product: protein A
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
- type: CDS
- locus tag: SAOUHSC_00069
- symbol: SAOUHSC_00069
- product: protein A
- replicon: chromosome
- strand: -
- coordinates: 73429..74979
- length: 1551
- essential: no DEG other strains
⊟Accession numbers[edit | edit source]
- Gene ID: 3919448 NCBI
- RefSeq: YP_498670 NCBI
- BioCyc: G1I0R-64 BioCyc
- MicrobesOnline: 1288564 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421
481
541
601
661
721
781
841
901
961
1021
1081
1141
1201
1261
1321
1381
1441
1501TTGAAAAAGAAAAACATTTATTCAATTCGTAAACTAGGTGTAGGTATTGCATCTGTAACT
TTAGGTACATTACTTATATCTGGTGGCGTAACACCTGCTGCAAATGCTGCGCAACACGAT
GAAGCTCAACAAAATGCTTTTTATCAAGTCTTAAATATGCCTAACTTAAATGCTGATCAA
CGCAATGGTTTTATCCAAAGCCTTAAAGATGATCCAAGCCAAAGTGCTAACGTTTTAGGT
GAAGCTCAAAAACTTAATGACTCTCAAGCTCCAAAAGCTGATGCGCAACAAAATAACTTC
AACAAAGATCAACAAAGCGCCTTCTATGAAATCTTGAACATGCCTAACTTAAACGAAGCG
CAACGTAACGGCTTCATTCAAAGTCTTAAAGACGACCCAAGCCAAAGCACTAACGTTTTA
GGTGAAGCTAAAAAATTAAACGAATCTCAAGCACCGAAAGCTGATAACAATTTCAACAAA
GAACAACAAAATGCTTTCTATGAAATCTTGAATATGCCTAACTTAAACGAAGAACAACGC
AATGGTTTCATCCAAAGCTTAAAAGATGACCCAAGCCAAAGTGCTAACCTATTGTCAGAA
GCTAAAAAGTTAAATGAATCTCAAGCACCGAAAGCGGATAACAAATTCAACAAAGAACAA
CAAAATGCTTTCTATGAAATCTTACATTTACCTAACTTAAACGAAGAACAACGCAATGGT
TTCATCCAAAGCCTAAAAGATGACCCAAGCCAAAGCGCTAACCTTTTAGCAGAAGCTAAA
AAGCTAAATGATGCTCAAGCACCAAAAGCTGACAACAAATTCAACAAAGAACAACAAAAT
GCTTTCTATGAAATTTTACATTTACCTAACTTAACTGAAGAACAACGTAACGGCTTCATC
CAAAGCCTTAAAGACGATCCTTCAGTGAGCAAAGAAATTTTAGCAGAAGCTAAAAAGCTA
AACGATGCTCAAGCACCAAAAGAGGAAGACAATAACAAGCCTGGCAAAGAAGACAATAAC
AAGCCTGGCAAAGAAGACAACAACAAGCCTGGTAAAGAAGACAACAACAAGCCTGGTAAA
GAAGACAACAACAAGCCTGGCAAAGAAGACGGCAACAAGCCTGGTAAAGAAGACAACAAA
AAACCTGGTAAAGAAGATGGCAACAAGCCTGGTAAAGAAGACAACAAAAAACCTGGTAAA
GAAGACGGCAACAAGCCTGGCAAAGAAGATGGCAACAAACCTGGTAAAGAAGATGGTAAC
GGAGTACATGTCGTTAAACCTGGTGATACAGTAAATGACATTGCAAAAGCAAACGGCACT
ACTGCTGACAAAATTGCTGCAGATAACAAATTAGCTGATAAAAACATGATCAAACCTGGT
CAAGAACTTGTTGTTGATAAGAAGCAACCAGCAAACCATGCAGATGCTAACAAAGCTCAA
GCATTACCAGAAACTGGTGAAGAAAATCCATTCATCGGTACAACTGTATTTGGTGGATTA
TCATTAGCCTTAGGTGCAGCGTTATTAGCTGGACGTCGTCGCGAACTATAA60
120
180
240
300
360
420
480
540
600
660
720
780
840
900
960
1020
1080
1140
1200
1260
1320
1380
1440
1500
1551
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SAOUHSC_00069
- symbol: SAOUHSC_00069
- description: protein A
- length: 516
- theoretical pI: 5.37468
- theoretical MW: 56436.8
- GRAVY: -1.16298
⊟Function[edit | edit source]
- TIGRFAM: gram-positive signal peptide, YSIRK family (TIGR01168; HMM-score: 34.5)and 3 moreCell envelope Other LPXTG cell wall anchor domain (TIGR01167; HMM-score: 19.4)Cellular processes Sporulation and germination spore coat assembly protein SafA (TIGR02899; HMM-score: 17.5)Biosynthesis of cofactors, prosthetic groups, and carriers Glutathione and analogs bacillithiol biosynthesis deacetylase BshB2 (TIGR04000; EC 3.-.-.-; HMM-score: 8.8)
- TheSEED :
- Protein A, von Willebrand factor binding protein Spa
- PFAM: B_GA (CL0598) B; B domain (PF02216; HMM-score: 474.5)and 21 moreGA; GA module (PF01468; HMM-score: 81.2)LysM (CL0187) LysM; LysM domain (PF01476; HMM-score: 50.2)no clan defined YSIRK_signal; YSIRK type signal peptide (PF04650; HMM-score: 39.8)Octapeptide; Octapeptide repeat (PF03373; HMM-score: 32.7)UFL1; E3 UFM1-protein ligase 1-like domain (PF23659; HMM-score: 31.9)Gram_pos_anchor; LPXTG cell wall anchor motif (PF00746; HMM-score: 31.8)VBS-like (CL0705) Talin_IBS2B; Talin IBS2B domain (PF21896; HMM-score: 22.5)Acetyltrans (CL0257) HAT1_C; Histone acetyltransferase type B catalytic subunit, C-terminal (PF21183; HMM-score: 21.3)no clan defined IFT43; Intraflagellar transport protein 43 (PF15305; HMM-score: 17.6)TerB (CL0414) TerB; Tellurite resistance protein TerB (PF05099; HMM-score: 17)no clan defined MRP-63; Mitochondrial ribosome protein 63 (PF14978; HMM-score: 17)Dim_A_B_barrel (CL0032) DUF3291; Domain of unknown function (DUF3291) (PF11695; HMM-score: 16.7)no clan defined CLAMP; Flagellar C1a complex subunit C1a-32 (PF14769; HMM-score: 15.5)SNARE-fusion (CL0445) V-SNARE; Vesicle transport v-SNARE protein N-terminus (PF05008; HMM-score: 15.4)LysM (CL0187) LysM3_LYK4_5; LYK4/5 LysM3 domain (PF23473; HMM-score: 14.3)E-set (CL0159) GH85_C; GH85 endohexosaminidases, C-terminal domain (PF21910; HMM-score: 13.3)post-AAA (CL0604) DNApol3-delta_C; DNA polymerase III, delta subunit, C terminal (PF09115; HMM-score: 10)no clan defined NTS_2; N-terminal segments of P. falciparum erythrocyte membrane protein (PF15448; HMM-score: 9.6)ANIS5_cation-bd; SXP/RAL-2 family protein Ani s 5-like, metal-binding domain (PF02520; HMM-score: 8.9)MMgT; Membrane magnesium transporter (PF10270; HMM-score: 8.3)Met_repress (CL0057) DUF1778; Protein of unknown function (DUF1778) (PF08681; HMM-score: 7)
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors:
- effectors:
⊟Localization[edit | edit source]
- PSORTb: Cellwall
- Cytoplasmic Score: 0
- Cytoplasmic Membrane Score: 0.01
- Cellwall Score: 9.98
- Extracellular Score: 0.01
- Internal Helices: 2
- DeepLocPro: Cell wall & surface
- Cytoplasmic Score: 0.0001
- Cytoplasmic Membrane Score: 0.039
- Cell wall & surface Score: 0.9084
- Extracellular Score: 0.0524
- LocateP: LPxTG Cell-wall anchored
- Prediction by SwissProt Classification: Cell Wall
- Pathway Prediction: Sec-(SPI)
- Intracellular possibility: 0
- Signal peptide possibility: 1
- N-terminally Anchored Score: -2
- Predicted Cleavage Site: found LPxTG motif :LPETG
- SignalP: Signal peptide SP(Sec/SPI) length 36 aa
- SP(Sec/SPI): 0.986537
- TAT(Tat/SPI): 0.008981
- LIPO(Sec/SPII): 0.003804
- Cleavage Site: CS pos: 36-37. ANA-AQ. Pr: 0.9494
- predicted transmembrane helices (TMHMM): 1
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MKKKNIYSIRKLGVGIASVTLGTLLISGGVTPAANAAQHDEAQQNAFYQVLNMPNLNADQRNGFIQSLKDDPSQSANVLGEAQKLNDSQAPKADAQQNNFNKDQQSAFYEILNMPNLNEAQRNGFIQSLKDDPSQSTNVLGEAKKLNESQAPKADNNFNKEQQNAFYEILNMPNLNEEQRNGFIQSLKDDPSQSANLLSEAKKLNESQAPKADNKFNKEQQNAFYEILHLPNLNEEQRNGFIQSLKDDPSQSANLLAEAKKLNDAQAPKADNKFNKEQQNAFYEILHLPNLTEEQRNGFIQSLKDDPSVSKEILAEAKKLNDAQAPKEEDNNKPGKEDNNKPGKEDNNKPGKEDNNKPGKEDNNKPGKEDGNKPGKEDNKKPGKEDGNKPGKEDNKKPGKEDGNKPGKEDGNKPGKEDGNGVHVVKPGDTVNDIAKANGTTADKIAADNKLADKNMIKPGQELVVDKKQPANHADANKAQALPETGEENPFIGTTVFGGLSLALGAALLAGRRREL
⊟Experimental data[edit | edit source]
- experimentally validated: PeptideAtlas [1] [2]
- protein localization: data available for COL
- quantitative data / protein copy number per cell:
- interaction partners:
SAOUHSC_02494 (rpsE) 30S ribosomal protein S5 [3] (data from MRSA252) SAOUHSC_00530 elongation factor Tu [3] (data from MRSA252) SAOUHSC_01794 glyceraldehyde 3-phosphate dehydrogenase 2 [3] (data from MRSA252) SAOUHSC_01819 hypothetical protein [3] (data from MRSA252)
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
- MicrobesOnline: no polycistronic organisation predicted
⊟Regulation[edit | edit source]
- regulators: CcpA* regulon, RNAIII/S871 regulon
CcpA* (TF) important in Carbon catabolism; RegPrecise RNAIII/S871 (sRNA) important in Virulence, Quorum sensing; [4]
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: [5]
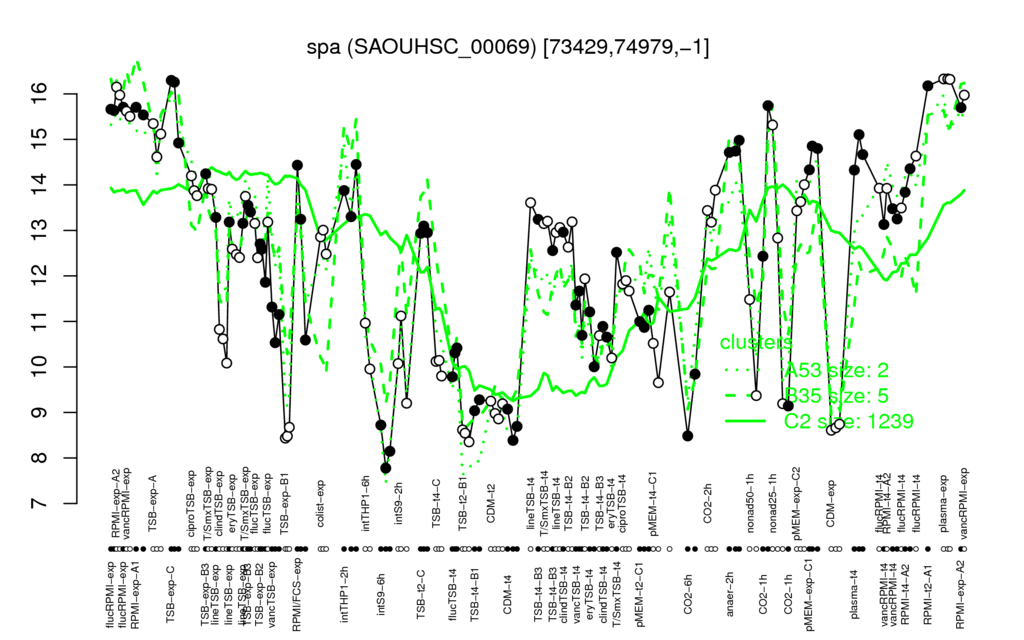 Multi-gene expression profiles
Multi-gene expression profiles
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
- Aureolib: no data available
⊟Protein stability[edit | edit source]
- half-life: no data available
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You can add further information about the gene and protein here. [edit]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ Maren Depke, Stephan Michalik, Alexander Rabe, Kristin Surmann, Lars Brinkmann, Nico Jehmlich, Jörg Bernhardt, Michael Hecker, Bernd Wollscheid, Zhi Sun, Robert L Moritz, Uwe Völker, Frank Schmidt
A peptide resource for the analysis of Staphylococcus aureus in host-pathogen interaction studies.
Proteomics: 2015, 15(21);3648-61
[PubMed:26224020] [WorldCat.org] [DOI] (I p) - ↑ Stephan Michalik, Maren Depke, Annette Murr, Manuela Gesell Salazar, Ulrike Kusebauch, Zhi Sun, Tanja C Meyer, Kristin Surmann, Henrike Pförtner, Petra Hildebrandt, Stefan Weiss, Laura Marcela Palma Medina, Melanie Gutjahr, Elke Hammer, Dörte Becher, Thomas Pribyl, Sven Hammerschmidt, Eric W Deutsch, Samuel L Bader, Michael Hecker, Robert L Moritz, Ulrike Mäder, Uwe Völker, Frank Schmidt
A global Staphylococcus aureus proteome resource applied to the in vivo characterization of host-pathogen interactions.
Sci Rep: 2017, 7(1);9718
[PubMed:28887440] [WorldCat.org] [DOI] (I e) - ↑ 3.0 3.1 3.2 3.3 Artem Cherkasov, Michael Hsing, Roya Zoraghi, Leonard J Foster, Raymond H See, Nikolay Stoynov, Jihong Jiang, Sukhbir Kaur, Tian Lian, Linda Jackson, Huansheng Gong, Rick Swayze, Emily Amandoron, Farhad Hormozdiari, Phuong Dao, Cenk Sahinalp, Osvaldo Santos-Filho, Peter Axerio-Cilies, Kendall Byler, William R McMaster, Robert C Brunham, B Brett Finlay, Neil E Reiner
Mapping the protein interaction network in methicillin-resistant Staphylococcus aureus.
J Proteome Res: 2011, 10(3);1139-50
[PubMed:21166474] [WorldCat.org] [DOI] (I p) - ↑ Delphine Bronesky, Zongfu Wu, Stefano Marzi, Philippe Walter, Thomas Geissmann, Karen Moreau, François Vandenesch, Isabelle Caldelari, Pascale Romby
Staphylococcus aureus RNAIII and Its Regulon Link Quorum Sensing, Stress Responses, Metabolic Adaptation, and Regulation of Virulence Gene Expression.
Annu Rev Microbiol: 2016, 70;299-316
[PubMed:27482744] [WorldCat.org] [DOI] (I p) - ↑ Ulrike Mäder, Pierre Nicolas, Maren Depke, Jan Pané-Farré, Michel Debarbouille, Magdalena M van der Kooi-Pol, Cyprien Guérin, Sandra Dérozier, Aurelia Hiron, Hanne Jarmer, Aurélie Leduc, Stephan Michalik, Ewoud Reilman, Marc Schaffer, Frank Schmidt, Philippe Bessières, Philippe Noirot, Michael Hecker, Tarek Msadek, Uwe Völker, Jan Maarten van Dijl
Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions.
PLoS Genet: 2016, 12(4);e1005962
[PubMed:27035918] [WorldCat.org] [DOI] (I e)
⊟Relevant publications[edit | edit source]
T J Foster, M Höök
Surface protein adhesins of Staphylococcus aureus.
Trends Microbiol: 1998, 6(12);484-8
[PubMed:10036727] [WorldCat.org] [DOI] (P p)M Graille, E A Stura, A L Corper, B J Sutton, M J Taussig, J B Charbonnier, G J Silverman
Crystal structure of a Staphylococcus aureus protein A domain complexed with the Fab fragment of a human IgM antibody: structural basis for recognition of B-cell receptors and superantigen activity.
Proc Natl Acad Sci U S A: 2000, 97(10);5399-404
[PubMed:10805799] [WorldCat.org] [DOI] (P p)Tanusree Das, Chhabinath Mandal, Chitra Mandal
Protein A--a new ligand for human C-reactive protein.
FEBS Lett: 2004, 576(1-2);107-13
[PubMed:15474020] [WorldCat.org] [DOI] (P p)Satoshi Sato, Tomasz L Religa, Alan R Fersht
Phi-analysis of the folding of the B domain of protein A using multiple optical probes.
J Mol Biol: 2006, 360(4);850-64
[PubMed:16782128] [WorldCat.org] [DOI] (P p)K Garbacz, L Piechowicz, J Galiński
[Characteristics of clumping factor- and protein A-negative methicillin-resistant Staphylococcus aureus identified in Poland]. [Caractéristiques des souches de Staphylococcus aureus facteur agglutinant- et protéine- A-négatifs résistants à la méthicilline isolées en Pologne.]
Med Mal Infect: 2006, 36(9);469-72
[PubMed:17011150] [WorldCat.org] [DOI] (P p)Marisa I Gómez, Maghnus O Seaghdha, Alice S Prince
Staphylococcus aureus protein A activates TACE through EGFR-dependent signaling.
EMBO J: 2007, 26(3);701-9
[PubMed:17255933] [WorldCat.org] [DOI] (P p)Andrea C DeDent, Molly McAdow, Olaf Schneewind
Distribution of protein A on the surface of Staphylococcus aureus.
J Bacteriol: 2007, 189(12);4473-84
[PubMed:17416657] [WorldCat.org] [DOI] (P p)F Huang, E Lerner, S Sato, D Amir, E Haas, A R Fersht
Time-resolved fluorescence resonance energy transfer study shows a compact denatured state of the B domain of protein A.
Biochemistry: 2009, 48(15);3468-76
[PubMed:19222162] [WorldCat.org] [DOI] (I p)Mark J J B Sibbald, Theresa Winter, Magdalena M van der Kooi-Pol, G Buist, E Tsompanidou, Tjibbe Bosma, Tina Schäfer, Knut Ohlsen, Michael Hecker, Haike Antelmann, Susanne Engelmann, Jan Maarten van Dijl
Synthetic effects of secG and secY2 mutations on exoproteome biogenesis in Staphylococcus aureus.
J Bacteriol: 2010, 192(14);3788-800
[PubMed:20472795] [WorldCat.org] [DOI] (I p)Hwan Keun Kim, Vilasack Thammavongsa, Olaf Schneewind, Dominique Missiakas
Recurrent infections and immune evasion strategies of Staphylococcus aureus.
Curr Opin Microbiol: 2012, 15(1);92-9
[PubMed:22088393] [WorldCat.org] [DOI] (I p)Samuel Becker, Matthew B Frankel, Olaf Schneewind, Dominique Missiakas
Release of protein A from the cell wall of Staphylococcus aureus.
Proc Natl Acad Sci U S A: 2014, 111(4);1574-9
[PubMed:24434550] [WorldCat.org] [DOI] (I p)M Uhlén, B Guss, B Nilsson, S Gatenbeck, L Philipson, M Lindberg
Complete sequence of the staphylococcal gene encoding protein A. A gene evolved through multiple duplications.
J Biol Chem: 1984, 259(3);1695-702
[PubMed:6319407] [WorldCat.org] (P p)S Löfdahl, B Guss, M Uhlén, L Philipson, M Lindberg
Gene for staphylococcal protein A.
Proc Natl Acad Sci U S A: 1983, 80(3);697-701
[PubMed:6338496] [WorldCat.org] [DOI] (P p)
