Jump to navigation
Jump to search
NCBI: 03-AUG-2016
⊟Summary[edit | edit source]
- organism: Staphylococcus aureus NCTC8325
- locus tag: SAOUHSC_00155
- pan locus tag?: SAUPAN001032000
- symbol: SAOUHSC_00155
- pan gene symbol?: glcA
- synonym:
- product: PTS system glucose-specific protein
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
- type: CDS
- locus tag: SAOUHSC_00155
- symbol: SAOUHSC_00155
- product: PTS system glucose-specific protein
- replicon: chromosome
- strand: -
- coordinates: 166663..168660
- length: 1998
- essential: no DEG other strains
⊟Accession numbers[edit | edit source]
- Gene ID: 3919863 NCBI
- RefSeq: YP_498754 NCBI
- BioCyc: G1I0R-145 BioCyc
- MicrobesOnline: 1288648 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421
481
541
601
661
721
781
841
901
961
1021
1081
1141
1201
1261
1321
1381
1441
1501
1561
1621
1681
1741
1801
1861
1921
1981ATGTTACCTGTTGCGATTTTACCAGCAGCTGGTCTGTTATTAGCTATCGGTACAGCTATG
CAAGGTGAATCATTACAACACTACTTGCCGTTTATACAAAATGGTGGCGTACAAACTGTC
GCTAAATTAATGACAGGTGCTGGTGGTATCATTTTTGATAACTTGCCTATGATTTTCGCA
TTAGGTGTCGCAATCGGATTAGCTGGCGGTGATGGCGTAGCAGCTATCGCAGCATTCGTC
GGTTACATAATCATGAACAAAACAATGGGCGACTTTTTACAAGTTACACCTAAGAATATT
GGTGATCCAGCGAGTGGTTACGCTAGCATTTTAGGTATCCCAACATTACAAACAGGTGTG
TTCGGCGGTATTATAATCGGGGCCCTGGCAGCTTGGTGTTATAACAAGTTCTATAACATT
AACTTACCATCTTATTTAGGTTTCTTCGCTGGTAAGCGTTTCGTACCTATTATGATGGCT
ACAACATCATTTATTTTAGCATTCCCAATGGCATTAATTTGGCCAACGATTCAATCAGGA
TTAAATGCATTCAGTACAGGATTATTAGATTCAAATACTGGTGTTGCCGTATTCTTATTT
GGTTTCATCAAGCGTTTATTAATTCCATTCGGTCTACATCACATTTTCCACGCACCGTTC
TGGTTCGAGTTTGGTTCATGGAAAAATGCAGCTGGTGAAATTATTCACGGTGACCAACGT
ATCTTTATCGAACAAATTCGTGAAGGCGCACATTTGACAGCTGGTAAATTCATGCAAGGT
GAATTCCCTGTTATGATGTTCGGTTTACCTGCAGCAGCTTTAGCAATTTATCACACAGCT
AAACCTGAAAATAAGAAAGTAGTAGCAGGTTTAATGGGTTCTGCTGCTTTAACATCATTC
TTAACTGGTATTACAGAACCATTAGAATTCTCATTCTTATTTGTAGCACCATTATTATTC
TTTATTCACGCAGTACTTGATGGTTTATCATTCTTAACATTGTACTTATTAGATCTTCAT
CTAGGTTATACATTCTCAGGTGGTTTCATCGACTACTTCTTACTCGGTATACTACCTAAT
AAGACACAATGGTGGTTAGTCATTCCTGTAGGTCTTGTATACGCAGTTATTTACTACTTC
GTATTCCGATTCTTAATTGTAAAATTAAAATACAAAACACCAGGTCGTGAAGATAAACAA
TCACAAGCGGCTACTGCTTCAGCAACTGAATTACCATATGCAGTATTAGAAGCTATGGGT
GGCAAAGCAAACATTAAACATTTAGACGCTTGTATCACACGTCTACGTGTTGAAGTTAAC
GACAAATCTAAAGTTGATGTTCCTGGTTTGAAAGATTTAGGCGCATCTGGTGTATTAGAA
GTCGGCAATAATATGCAAGCAATTTTTGGTCCTAAATCTGACCAAATCAAACATGAAATG
CAACAGATTATGAATGGTCAAGTAGTAGAAAATCCTACTACTATGGAAGACGATAAAGAC
GAAACTGTTGTTGTTGCAGAAGATAAATCTGCAACAAGCGAATTGAGCCATATCGTGCAT
GCACCATTAACTGGTGAAGTAACACCATTATCAGAAGTGCCTGATCAAGTGTTCAGCGAA
AAAATGATGGGTGACGGTATCGCTATCAAACCTTCACAAGGTGAAGTTCGTGCACCATTC
AACGGTAAAGTACAAATGATTTTCCCAACAAAACATGCAATTGGTCTTGTATCAGATAGT
GGTTTAGAACTATTAATCCACATCGGTTTAGACACTGTTAAATTAAACGGAGAAGGCTTT
ACTTTACATGTTGAGGAAGGTCAAGAAGTTAAACAAGGTGATTTATTAATCAACTTTGAT
TTAGACTACATCCGCAATCATGCAAAGAGTGATATTACGCCTATTATCGTGACACAAGGA
AACATTACAAACCTTGATTTTAAACAAGGTGAACATGGCAACATTTCATTTGGCGATCAA
TTATTTGAAGCTAAATAA60
120
180
240
300
360
420
480
540
600
660
720
780
840
900
960
1020
1080
1140
1200
1260
1320
1380
1440
1500
1560
1620
1680
1740
1800
1860
1920
1980
1998
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SAOUHSC_00155
- symbol: SAOUHSC_00155
- description: PTS system glucose-specific protein
- length: 665
- theoretical pI: 5.7965
- theoretical MW: 72055.3
- GRAVY: 0.336241
⊟Function[edit | edit source]
- reaction: EC 2.7.1.69? ExPASyTransferred entry: 2.7.1.191, 2.7.1.192, 2.7.1.193, 2.7.1.194, 2.7.1.195, 2.7.1.196, 2.7.1.197, 2.7.1.198, 2.7.1.199, 2.7.1.200, 2.7.1.201, 2.7.1.202, 2.7.1.203, 2.7.1.204, 2.7.1.205, 2.7.1.206, 2.7.1.207 and 2.7.1.208EC 2.7.1.199? ExPASyProtein-N(pi)-phosphohistidine--D-glucose phosphotransferase [Protein]-N(pi)-phospho-L-histidine + D-glucose(Side 1) = [protein]-L-histidine + D-glucose 6-phosphate(Side 2)
- TIGRFAM: Transport and binding proteins Carbohydrates, organic alcohols, and acids PTS system, glucose-specific IIBC component (TIGR02002; EC 2.7.1.69; HMM-score: 771.6)and 15 moreTransport and binding proteins Carbohydrates, organic alcohols, and acids PTS system, N-acetylglucosamine-specific IIBC component (TIGR01998; EC 2.7.1.69; HMM-score: 460.5)Transport and binding proteins Carbohydrates, organic alcohols, and acids PTS system, maltose and glucose-specific IIBC component (TIGR02004; EC 2.7.1.69; HMM-score: 385.3)Transport and binding proteins Carbohydrates, organic alcohols, and acids PTS system, maltose and glucose-specific subfamily, IIC component (TIGR00852; HMM-score: 279.3)Signal transduction PTS PTS system, maltose and glucose-specific subfamily, IIC component (TIGR00852; HMM-score: 279.3)Transport and binding proteins Carbohydrates, organic alcohols, and acids PTS system, alpha-glucoside-specific IIBC component (TIGR02005; EC 2.7.1.69; HMM-score: 271)Transport and binding proteins Carbohydrates, organic alcohols, and acids PTS system, IIBC component (TIGR02003; EC 2.7.1.69; HMM-score: 230.2)PTS system, beta-glucoside-specific IIABC component (TIGR01995; EC 2.7.1.69; HMM-score: 209.8)Transport and binding proteins Carbohydrates, organic alcohols, and acids PTS system, glucose subfamily, IIA component (TIGR00830; HMM-score: 165.5)Signal transduction PTS PTS system, glucose subfamily, IIA component (TIGR00830; HMM-score: 165.5)Transport and binding proteins Carbohydrates, organic alcohols, and acids PTS system, glucose-like IIB component (TIGR00826; EC 2.7.1.69; HMM-score: 73.8)Signal transduction PTS PTS system, glucose-like IIB component (TIGR00826; EC 2.7.1.69; HMM-score: 73.8)PTS system, sucrose-specific IIBC component (TIGR01996; EC 2.7.1.69; HMM-score: 55.9)PTS system, trehalose-specific IIBC component (TIGR01992; EC 2.7.1.69; HMM-score: 28.5)Protein fate Protein and peptide secretion and trafficking type I secretion membrane fusion protein, HlyD family (TIGR01843; HMM-score: 16.6)Transport and binding proteins Unknown substrate efflux transporter, RND family, MFP subunit (TIGR01730; HMM-score: 14.3)
- TheSEED :
- PTS system, glucose-specific IIA component
- PTS system, glucose-specific IIB component (EC 2.7.1.69)
- PTS system, glucose-specific IIC component
Carbohydrates Di- and oligosaccharides Trehalose Uptake and Utilization PTS system, glucose-specific IIA componentand 2 more - PFAM: PTS_EIIC (CL0493) PTS_EIIC; Phosphotransferase system, EIIC (PF02378; HMM-score: 315.4)and 6 moreHybrid (CL0105) PTS_EIIA_1; phosphoenolpyruvate-dependent sugar phosphotransferase system, EIIA 1 (PF00358; HMM-score: 183.1)no clan defined PTS_EIIB; phosphotransferase system, EIIB (PF00367; HMM-score: 57.1)Hybrid (CL0105) Biotin_lipoyl_2; Biotin-lipoyl like (PF13533; HMM-score: 20.9)no clan defined PIG-P; PIG-P (PF08510; HMM-score: 14.1)Hybrid (CL0105) Peptidase_M23; Peptidase family M23 (PF01551; HMM-score: 12.5)no clan defined RDD; RDD family (PF06271; HMM-score: 9.5)
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors:
- effectors:
⊟Localization[edit | edit source]
- PSORTb: Cytoplasmic Membrane
- Cytoplasmic Score: 0
- Cytoplasmic Membrane Score: 10
- Cellwall Score: 0
- Extracellular Score: 0
- Internal Helices: 10
- DeepLocPro: Cytoplasmic Membrane
- Cytoplasmic Score: 0.0002
- Cytoplasmic Membrane Score: 0.9991
- Cell wall & surface Score: 0
- Extracellular Score: 0.0006
- LocateP: Multi-transmembrane
- Prediction by SwissProt Classification: Membrane
- Pathway Prediction: Sec-(SPI)
- Intracellular possibility: 0
- Signal peptide possibility: 0
- N-terminally Anchored Score: 1
- Predicted Cleavage Site: No CleavageSite
- SignalP: no predicted signal peptide
- SP(Sec/SPI): 0.26208
- TAT(Tat/SPI): 0.003
- LIPO(Sec/SPII): 0.149366
- predicted transmembrane helices (TMHMM): 10
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MLPVAILPAAGLLLAIGTAMQGESLQHYLPFIQNGGVQTVAKLMTGAGGIIFDNLPMIFALGVAIGLAGGDGVAAIAAFVGYIIMNKTMGDFLQVTPKNIGDPASGYASILGIPTLQTGVFGGIIIGALAAWCYNKFYNINLPSYLGFFAGKRFVPIMMATTSFILAFPMALIWPTIQSGLNAFSTGLLDSNTGVAVFLFGFIKRLLIPFGLHHIFHAPFWFEFGSWKNAAGEIIHGDQRIFIEQIREGAHLTAGKFMQGEFPVMMFGLPAAALAIYHTAKPENKKVVAGLMGSAALTSFLTGITEPLEFSFLFVAPLLFFIHAVLDGLSFLTLYLLDLHLGYTFSGGFIDYFLLGILPNKTQWWLVIPVGLVYAVIYYFVFRFLIVKLKYKTPGREDKQSQAATASATELPYAVLEAMGGKANIKHLDACITRLRVEVNDKSKVDVPGLKDLGASGVLEVGNNMQAIFGPKSDQIKHEMQQIMNGQVVENPTTMEDDKDETVVVAEDKSATSELSHIVHAPLTGEVTPLSEVPDQVFSEKMMGDGIAIKPSQGEVRAPFNGKVQMIFPTKHAIGLVSDSGLELLIHIGLDTVKLNGEGFTLHVEEGQEVKQGDLLINFDLDYIRNHAKSDITPIIVTQGNITNLDFKQGEHGNISFGDQLFEAK
⊟Experimental data[edit | edit source]
- experimentally validated: PeptideAtlas [1] [2]
- protein localization: data available for COL
- quantitative data / protein copy number per cell:
- interaction partners:
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
- predicted SigA promoter [3] : SAOUHSC_00155 < S51
⊟Regulation[edit | edit source]
- regulator: CcpA* regulon
CcpA* (TF) important in Carbon catabolism; RegPrecise
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: [3]
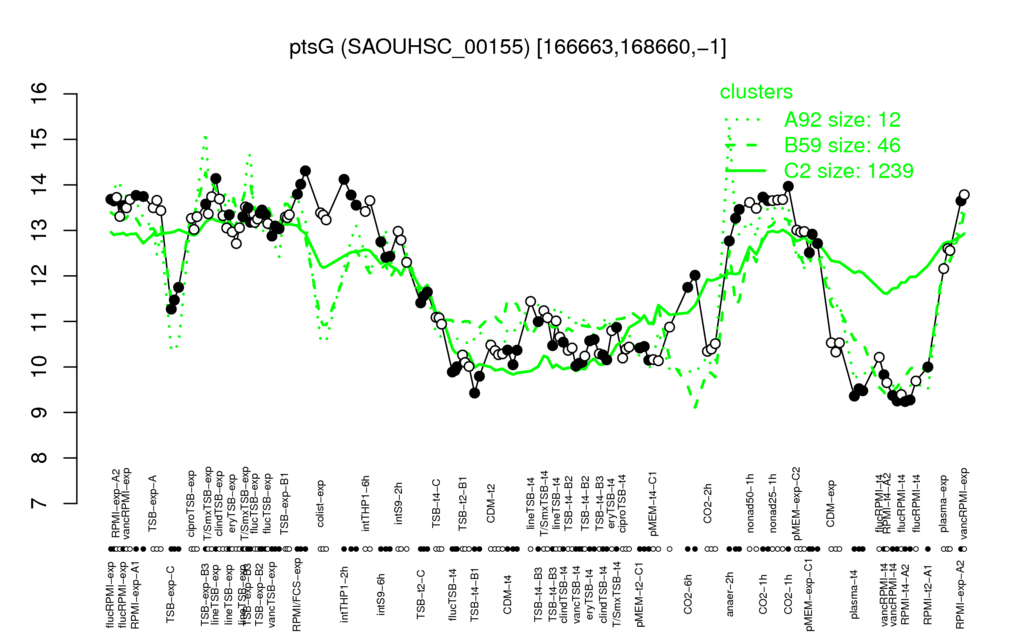 Multi-gene expression profiles
Multi-gene expression profiles
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
- Aureolib: no data available
⊟Protein stability[edit | edit source]
- half-life: no data available
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You can add further information about the gene and protein here. [edit]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ Maren Depke, Stephan Michalik, Alexander Rabe, Kristin Surmann, Lars Brinkmann, Nico Jehmlich, Jörg Bernhardt, Michael Hecker, Bernd Wollscheid, Zhi Sun, Robert L Moritz, Uwe Völker, Frank Schmidt
A peptide resource for the analysis of Staphylococcus aureus in host-pathogen interaction studies.
Proteomics: 2015, 15(21);3648-61
[PubMed:26224020] [WorldCat.org] [DOI] (I p) - ↑ Stephan Michalik, Maren Depke, Annette Murr, Manuela Gesell Salazar, Ulrike Kusebauch, Zhi Sun, Tanja C Meyer, Kristin Surmann, Henrike Pförtner, Petra Hildebrandt, Stefan Weiss, Laura Marcela Palma Medina, Melanie Gutjahr, Elke Hammer, Dörte Becher, Thomas Pribyl, Sven Hammerschmidt, Eric W Deutsch, Samuel L Bader, Michael Hecker, Robert L Moritz, Ulrike Mäder, Uwe Völker, Frank Schmidt
A global Staphylococcus aureus proteome resource applied to the in vivo characterization of host-pathogen interactions.
Sci Rep: 2017, 7(1);9718
[PubMed:28887440] [WorldCat.org] [DOI] (I e) - ↑ 3.0 3.1 Ulrike Mäder, Pierre Nicolas, Maren Depke, Jan Pané-Farré, Michel Debarbouille, Magdalena M van der Kooi-Pol, Cyprien Guérin, Sandra Dérozier, Aurelia Hiron, Hanne Jarmer, Aurélie Leduc, Stephan Michalik, Ewoud Reilman, Marc Schaffer, Frank Schmidt, Philippe Bessières, Philippe Noirot, Michael Hecker, Tarek Msadek, Uwe Völker, Jan Maarten van Dijl
Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions.
PLoS Genet: 2016, 12(4);e1005962
[PubMed:27035918] [WorldCat.org] [DOI] (I e)
