Jump to navigation
Jump to search
NCBI: 03-AUG-2016
⊟Summary[edit | edit source]
- organism: Staphylococcus aureus NCTC8325
- locus tag: SAOUHSC_00882
- pan locus tag?: SAUPAN003046000
- symbol: SAOUHSC_00882
- pan gene symbol?: —
- synonym:
- product: hypothetical protein
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
- type: CDS
- locus tag: SAOUHSC_00882
- symbol: SAOUHSC_00882
- product: hypothetical protein
- replicon: chromosome
- strand: -
- coordinates: 846790..847944
- length: 1155
- essential: no DEG other strains
⊟Accession numbers[edit | edit source]
- Gene ID: 3919229 NCBI
- RefSeq: YP_499435 NCBI
- BioCyc: G1I0R-825 BioCyc
- MicrobesOnline: 1289346 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421
481
541
601
661
721
781
841
901
961
1021
1081
1141ATGCATTGGACAATTATCGGCGGTGGCATACAGGGAACTGCAATCGCACAAAAACTATTA
TCAAGCGGATTAACAACAGACCGATTAACAATCATTGACCCACACGAAACTTTTTGCCAA
AGGTTTAACTCATATACAAATCGAATAGAAATGCCTTATTTAAGATCACCGATTGTACAT
CACGTACATCCACAACCGTTCCATCTAAAACAATTCGCTAAACAGCACCAATATACAAAT
GCTTTTTATGGTCCTTATCAACGACCTGAATTGACAATGTTTATGGATCATATTGCACAT
GCTTCTAAACAATATCAATTAGAGGATTGCTTGGTTCAAGGTTTAGTTCAAACTTTAGAT
AAACAAGAAGACAAATGGCATATCAAGTTAGAAGATGGACAAATTATCACTACAGATTGC
GTCGTTATTGCAATAGGCAGTACAAATATTCCGTTTATGCCTGACATTTTAAAAGACAAA
CAGAATGTAAATCATATCTTCGAGAAAGAACTTGATCAAGTAGTATATGATAAGACCGAT
CATATCGTTGGTAGCGGCATTACTGCTGCACATCTTGCACTTAAATTGTTAAATCATGAT
AACGATAAAAAGATTCATTTATGGCTAAATAAAGATATTGAAATACATGACTTTGATGCT
GATCCTGGTTGGTTAGGTCCGAAAAATATGTCTTCATTTTTAAGTACTAAAAGCATGCCT
GAAAGAAATGCCATTGTACAACGCGAACGTCATAAAGGATCAATGCCTCACGAACTGTAC
TTACGCCTTAAAAAACATATTAAAAATGGTCGTATAAATGTGCATAAAACACCTATCACT
CAAATTAGTGGTGGTGTAATTAACACTGAAAATGATTCTGTTCCATATCAACAGATTATG
GTTGCAACTGGTTTTGAACAAGATTTTATGTCACAACCACTTATTAAGCAATTAATACAA
AATTATGATGCACCTATCAACGAATGTAATTACCCTGTTATTTCCGAAAAATTAGAATGG
ATACCAAATCTATTTGTCGCAGGATGCTTTGCAGACTTAGAATTAGGACCATTTGGTAGA
AATGTTATGGGTGGCCGTAAAGCTGCCGAACGCATTGAACAAGCATTTCTAAAACTACAA
CAATATAGCGCATAA60
120
180
240
300
360
420
480
540
600
660
720
780
840
900
960
1020
1080
1140
1155
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SAOUHSC_00882
- symbol: SAOUHSC_00882
- description: hypothetical protein
- length: 384
- theoretical pI: 7.07664
- theoretical MW: 43955
- GRAVY: -0.403906
⊟Function[edit | edit source]
- TIGRFAM: Protein synthesis tRNA and rRNA base modification tRNA U-34 5-methylaminomethyl-2-thiouridine biosynthesis protein MnmC, C-terminal domain (TIGR03197; HMM-score: 30.1)Biosynthesis of cofactors, prosthetic groups, and carriers Menaquinone and ubiquinone ubiquinone biosynthesis hydroxylase, UbiH/UbiF/VisC/COQ6 family (TIGR01988; EC 1.14.13.-; HMM-score: 25.3)and 9 morelycopene cyclase family protein (TIGR01790; HMM-score: 21.5)putative histamine N-monooxygenase (TIGR04439; EC 1.14.13.-; HMM-score: 16.3)mycothione reductase (TIGR03452; EC 1.8.1.15; HMM-score: 13.8)Biosynthesis of cofactors, prosthetic groups, and carriers Menaquinone and ubiquinone 2-polyprenyl-6-methoxyphenol 4-hydroxylase (TIGR01984; EC 1.14.13.-; HMM-score: 13.4)Biosynthesis of cofactors, prosthetic groups, and carriers Thiamine glycine oxidase ThiO (TIGR02352; EC 1.4.3.19; HMM-score: 12.3)Cellular processes Detoxification alkyl hydroperoxide reductase subunit F (TIGR03140; EC 1.8.1.-; HMM-score: 12.1)Cellular processes Adaptations to atypical conditions alkyl hydroperoxide reductase subunit F (TIGR03140; EC 1.8.1.-; HMM-score: 12.1)Unknown function Enzymes of unknown specificity flavoprotein, HI0933 family (TIGR00275; HMM-score: 11.7)Cellular processes Biosynthesis of natural products 2,3-diaminopropionate biosynthesis protein SbnB (TIGR03944; HMM-score: 11)
- TheSEED :
- FIG01108091: hypothetical protein
- PFAM: NADP_Rossmann (CL0063) NAD_binding_9; FAD-NAD(P)-binding (PF13454; HMM-score: 45.6)Pyr_redox_2; Pyridine nucleotide-disulphide oxidoreductase (PF07992; HMM-score: 40.4)Pyr_redox_3; Pyridine nucleotide-disulphide oxidoreductase (PF13738; HMM-score: 40)and 9 moreNAD_binding_8; NAD(P)-binding Rossmann-like domain (PF13450; HMM-score: 25.3)Lys_Orn_oxgnase; L-lysine 6-monooxygenase/L-ornithine 5-monooxygenase (PF13434; HMM-score: 23)no clan defined MptE-like; 6-hydroxymethylpterin diphosphokinase MptE-like (PF01973; HMM-score: 21.2)NADP_Rossmann (CL0063) Sacchrp_dh_NADP; Saccharopine dehydrogenase NADP binding domain (PF03435; HMM-score: 18.5)DAO; FAD dependent oxidoreductase (PF01266; HMM-score: 17.5)F420_oxidored; NADP oxidoreductase coenzyme F420-dependent (PF03807; HMM-score: 16.6)Pyr_redox; Pyridine nucleotide-disulphide oxidoreductase (PF00070; HMM-score: 13.4)TrkA_N; TrkA-N domain (PF02254; HMM-score: 12.8)GIDA; Glucose inhibited division protein A (PF01134; HMM-score: 11.2)
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors:
- effectors:
⊟Localization[edit | edit source]
- PSORTb: Cytoplasmic
- Cytoplasmic Score: 7.5
- Cytoplasmic Membrane Score: 1.15
- Cellwall Score: 0.62
- Extracellular Score: 0.73
- Internal Helices: 0
- DeepLocPro: Cytoplasmic
- Cytoplasmic Score: 0.7528
- Cytoplasmic Membrane Score: 0.2289
- Cell wall & surface Score: 0.0002
- Extracellular Score: 0.0181
- LocateP: Intracellular
- Prediction by SwissProt Classification: Cytoplasmic
- Pathway Prediction: No pathway
- Intracellular possibility: 1
- Signal peptide possibility: -1
- N-terminally Anchored Score: 1
- Predicted Cleavage Site: No CleavageSite
- SignalP: no predicted signal peptide
- SP(Sec/SPI): 0.02305
- TAT(Tat/SPI): 0.002147
- LIPO(Sec/SPII): 0.015907
- predicted transmembrane helices (TMHMM): 0
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MHWTIIGGGIQGTAIAQKLLSSGLTTDRLTIIDPHETFCQRFNSYTNRIEMPYLRSPIVHHVHPQPFHLKQFAKQHQYTNAFYGPYQRPELTMFMDHIAHASKQYQLEDCLVQGLVQTLDKQEDKWHIKLEDGQIITTDCVVIAIGSTNIPFMPDILKDKQNVNHIFEKELDQVVYDKTDHIVGSGITAAHLALKLLNHDNDKKIHLWLNKDIEIHDFDADPGWLGPKNMSSFLSTKSMPERNAIVQRERHKGSMPHELYLRLKKHIKNGRINVHKTPITQISGGVINTENDSVPYQQIMVATGFEQDFMSQPLIKQLIQNYDAPINECNYPVISEKLEWIPNLFVAGCFADLELGPFGRNVMGGRKAAERIEQAFLKLQQYSA
⊟Experimental data[edit | edit source]
- experimentally validated:
- protein localization:
- quantitative data / protein copy number per cell:
- interaction partners:
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
- MicrobesOnline: no polycistronic organisation predicted
⊟Regulation[edit | edit source]
- regulators: GraR* (repression) regulon, Zur* (repression) regulon
GraR* (response regulator) important in cationic antimicrobial peptide (CAMP) resistance; [1] Zur* (TF) important in Zinc homeostasis; RegPrecise
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: [2]
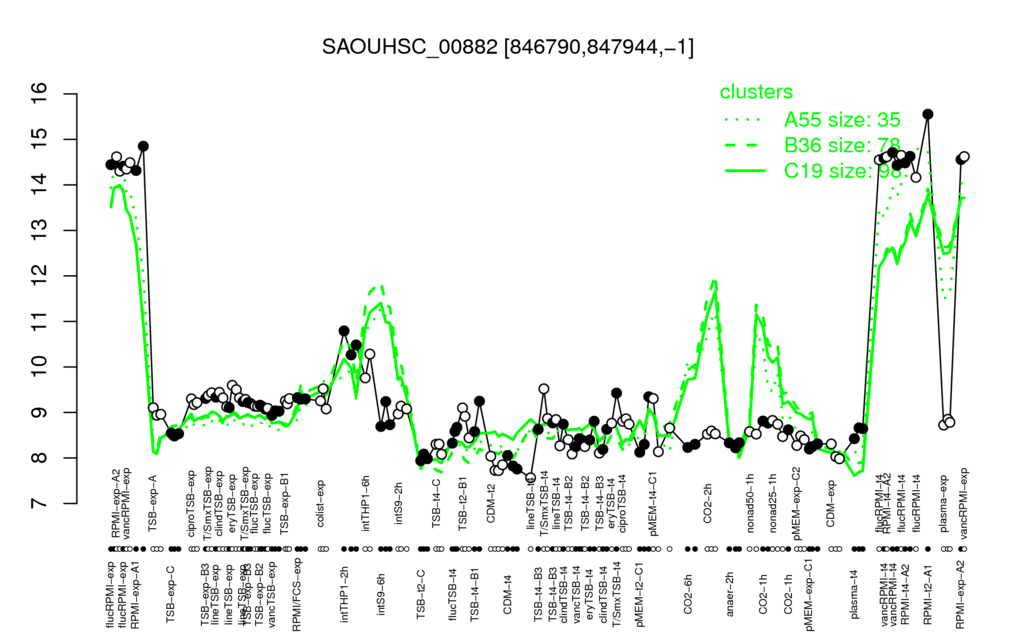 Multi-gene expression profiles
Multi-gene expression profiles
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
- Aureolib: no data available
⊟Protein stability[edit | edit source]
- half-life: no data available
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You can add further information about the gene and protein here. [edit]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ Mélanie Falord, Ulrike Mäder, Aurélia Hiron, Michel Débarbouillé, Tarek Msadek
Investigation of the Staphylococcus aureus GraSR regulon reveals novel links to virulence, stress response and cell wall signal transduction pathways.
PLoS One: 2011, 6(7);e21323
[PubMed:21765893] [WorldCat.org] [DOI] (I p) - ↑ Ulrike Mäder, Pierre Nicolas, Maren Depke, Jan Pané-Farré, Michel Debarbouille, Magdalena M van der Kooi-Pol, Cyprien Guérin, Sandra Dérozier, Aurelia Hiron, Hanne Jarmer, Aurélie Leduc, Stephan Michalik, Ewoud Reilman, Marc Schaffer, Frank Schmidt, Philippe Bessières, Philippe Noirot, Michael Hecker, Tarek Msadek, Uwe Völker, Jan Maarten van Dijl
Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions.
PLoS Genet: 2016, 12(4);e1005962
[PubMed:27035918] [WorldCat.org] [DOI] (I e)
