Jump to navigation
Jump to search
NCBI: 03-AUG-2016
⊟Summary[edit | edit source]
- organism: Staphylococcus aureus NCTC8325
- locus tag: SAOUHSC_00980
- pan locus tag?: SAUPAN003242000
- symbol: SAOUHSC_00980
- pan gene symbol?: menA
- synonym:
- product: 1,4-dihydroxy-2-naphthoate octaprenyltransferase
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
⊟Accession numbers[edit | edit source]
- Gene ID: 3920125 NCBI
- RefSeq: YP_499533 NCBI
- BioCyc: G1I0R-923 BioCyc
- MicrobesOnline: 1289446 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421
481
541
601
661
721
781
841
901ATGAGTAATCAATATCAGCAATATTCTACAGTTAAGAAATATTGGCATTTAATGCGTCCT
CATACATTAACTGCTTCCGTAGTACCCGTTTTAGTTGGTACAGCAGCATCTAAAATATAT
TTTCTTGGTAGCGAAGATCATATTAAAATCAGCCTATTCATTGCCATGTTACTAGCATGC
TTACTTATTCAAGCAGCAACTAATATGTTTAATGAATACTATGATTATAAAAAAGGCCTC
GATGATCATGAATCTGTAGGCATTGGTGGTGCCATTGTTCGCAACGGTATGAGCCCAGAG
CTTGTGCTACGATTAGCCATTGCATTTTACATCTTAGCAGCAATATTAGGTTTGTTTTTA
GCTGCTAACTCTTCATTTTGGTTATTACCAGTTGGATTAGTATGTATGGCTGTTGGTTAC
CTATATACAGGTGGCCCTTTCCCTATTTCATGGACGCCTTTCGGTGAATTATTCTCAGGC
GTATTTATGGGTATGTTTATTATCGTTATTGCATTCTTTATTCAAACTGGCAATATTCAA
AGTTATGTAATTTGGTTAAGTGTACCTATAGTAATCACTATCGGTTTAATTAATATGGCT
AACAATATTCGCGACCGTGTCAAAGATAAAGCAAGTGGTCGCAAAACTTTACCCATTCTA
TTAGGTAAAAATGCTTCTTTAACATTTATGGCAATCATGTACTTTATCGCTTATGCCTTT
ATTGTACTTACGATCATTATTAAACCTGGTGGCTCATTATTTTACTTACTTGCGTTGTTA
TCATTCCCAATGCCTGTTAAAGTTATCAGACGTTTCAAGAAGAATGATACACCGCCTACA
ATGATGCCAGCAATGGCTGCTGCTGGTAAAACAAATACATTTTTCGGTTTATTATATGCA
TTAGGTATTTATATTAGTGCATTATTTGCAGGCATTTAA60
120
180
240
300
360
420
480
540
600
660
720
780
840
900
939
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SAOUHSC_00980
- symbol: SAOUHSC_00980
- description: 1,4-dihydroxy-2-naphthoate octaprenyltransferase
- length: 312
- theoretical pI: 9.91895
- theoretical MW: 34400
- GRAVY: 0.733654
⊟Function[edit | edit source]
- reaction: EC 2.5.1.74? ExPASy1,4-dihydroxy-2-naphthoate polyprenyltransferase An all-trans-polyprenyl diphosphate + 1,4-dihydroxy-2-naphthoate = a demethylmenaquinol + diphosphate + CO2
- TIGRFAM: Biosynthesis of cofactors, prosthetic groups, and carriers Menaquinone and ubiquinone 1,4-dihydroxy-2-naphthoate octaprenyltransferase (TIGR00751; EC 2.5.1.74; HMM-score: 99.2)Biosynthesis of cofactors, prosthetic groups, and carriers Menaquinone and ubiquinone 1,4-dihydroxy-2-naphthoate phytyltransferase (TIGR02235; EC 2.5.1.-; HMM-score: 98.1)and 2 moreBiosynthesis of cofactors, prosthetic groups, and carriers Chlorophyll and bacteriochlorphyll chlorophyll synthase ChlG (TIGR02056; EC 2.5.1.62; HMM-score: 23.7)Biosynthesis of cofactors, prosthetic groups, and carriers Menaquinone and ubiquinone putative 4-hydroxybenzoate polyprenyltransferase (TIGR01475; EC 2.5.1.-; HMM-score: 21.5)
- TheSEED :
- 1,4-dihydroxy-2-naphthoate polyprenyltransferase (EC 2.5.1.74)
Cofactors, Vitamins, Prosthetic Groups, Pigments Quinone cofactors Menaquinone and Phylloquinone Biosynthesis 1,4-dihydroxy-2-naphthoate polyprenyltransferase (EC 2.5.1.74)and 1 more - PFAM: Terp_synthase (CL0613) UbiA; UbiA prenyltransferase family (PF01040; HMM-score: 119.5)and 2 moreDMT (CL0184) DUF5989; Family of unknown function (DUF5989) (PF19451; HMM-score: 15.3)no clan defined DAGK_prokar; Prokaryotic diacylglycerol kinase (PF01219; HMM-score: 6.8)
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors:
- effectors:
⊟Localization[edit | edit source]
- PSORTb: Cytoplasmic Membrane
- Cytoplasmic Score: 0
- Cytoplasmic Membrane Score: 10
- Cellwall Score: 0
- Extracellular Score: 0
- Internal Helices: 9
- DeepLocPro: Cytoplasmic Membrane
- Cytoplasmic Score: 0
- Cytoplasmic Membrane Score: 0.9994
- Cell wall & surface Score: 0
- Extracellular Score: 0.0006
- LocateP: Multi-transmembrane
- Prediction by SwissProt Classification: Membrane
- Pathway Prediction: Sec-(SPI)
- Intracellular possibility: 0.17
- Signal peptide possibility: -0.5
- N-terminally Anchored Score: -1
- Predicted Cleavage Site: No CleavageSite
- SignalP: no predicted signal peptide
- SP(Sec/SPI): 0.010098
- TAT(Tat/SPI): 0.000713
- LIPO(Sec/SPII): 0.016984
- predicted transmembrane helices (TMHMM): 9
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MSNQYQQYSTVKKYWHLMRPHTLTASVVPVLVGTAASKIYFLGSEDHIKISLFIAMLLACLLIQAATNMFNEYYDYKKGLDDHESVGIGGAIVRNGMSPELVLRLAIAFYILAAILGLFLAANSSFWLLPVGLVCMAVGYLYTGGPFPISWTPFGELFSGVFMGMFIIVIAFFIQTGNIQSYVIWLSVPIVITIGLINMANNIRDRVKDKASGRKTLPILLGKNASLTFMAIMYFIAYAFIVLTIIIKPGGSLFYLLALLSFPMPVKVIRRFKKNDTPPTMMPAMAAAGKTNTFFGLLYALGIYISALFAGI
⊟Experimental data[edit | edit source]
- experimentally validated: PeptideAtlas [2] [3]
- protein localization: data available for COL
- quantitative data / protein copy number per cell:
- interaction partners:
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
- MicrobesOnline: no polycistronic organisation predicted
⊟Regulation[edit | edit source]
- regulator:
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: [4]
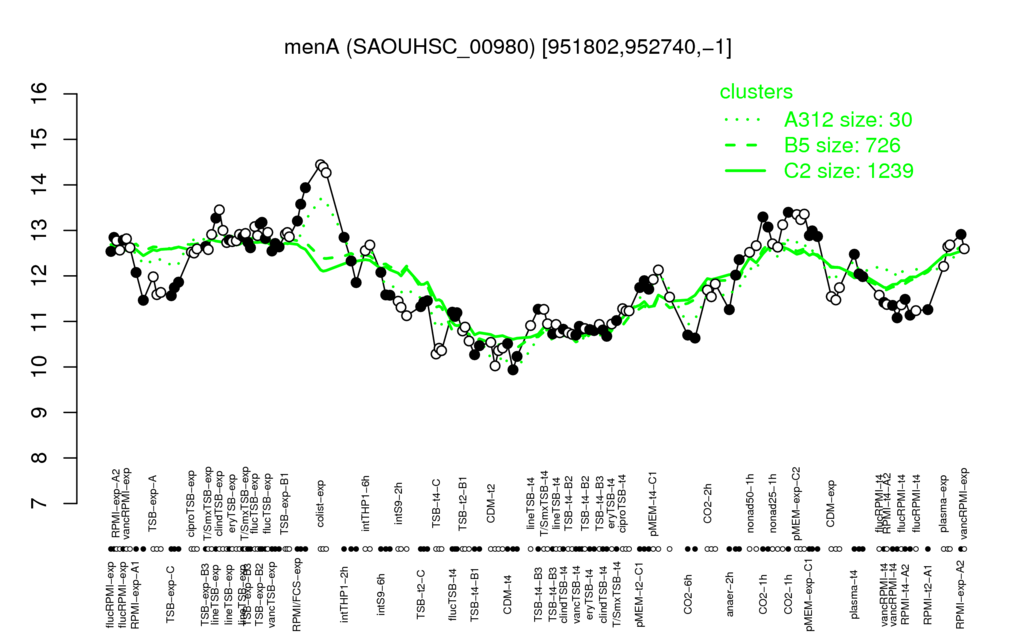 Multi-gene expression profiles
Multi-gene expression profiles
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
- Aureolib: no data available
⊟Protein stability[edit | edit source]
- half-life: no data available
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You can add further information about the gene and protein here. [edit]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ Roy R Chaudhuri, Andrew G Allen, Paul J Owen, Gil Shalom, Karl Stone, Marcus Harrison, Timothy A Burgis, Michael Lockyer, Jorge Garcia-Lara, Simon J Foster, Stephen J Pleasance, Sarah E Peters, Duncan J Maskell, Ian G Charles
Comprehensive identification of essential Staphylococcus aureus genes using Transposon-Mediated Differential Hybridisation (TMDH).
BMC Genomics: 2009, 10;291
[PubMed:19570206] [WorldCat.org] [DOI] (I e) - ↑ Maren Depke, Stephan Michalik, Alexander Rabe, Kristin Surmann, Lars Brinkmann, Nico Jehmlich, Jörg Bernhardt, Michael Hecker, Bernd Wollscheid, Zhi Sun, Robert L Moritz, Uwe Völker, Frank Schmidt
A peptide resource for the analysis of Staphylococcus aureus in host-pathogen interaction studies.
Proteomics: 2015, 15(21);3648-61
[PubMed:26224020] [WorldCat.org] [DOI] (I p) - ↑ Stephan Michalik, Maren Depke, Annette Murr, Manuela Gesell Salazar, Ulrike Kusebauch, Zhi Sun, Tanja C Meyer, Kristin Surmann, Henrike Pförtner, Petra Hildebrandt, Stefan Weiss, Laura Marcela Palma Medina, Melanie Gutjahr, Elke Hammer, Dörte Becher, Thomas Pribyl, Sven Hammerschmidt, Eric W Deutsch, Samuel L Bader, Michael Hecker, Robert L Moritz, Ulrike Mäder, Uwe Völker, Frank Schmidt
A global Staphylococcus aureus proteome resource applied to the in vivo characterization of host-pathogen interactions.
Sci Rep: 2017, 7(1);9718
[PubMed:28887440] [WorldCat.org] [DOI] (I e) - ↑ Ulrike Mäder, Pierre Nicolas, Maren Depke, Jan Pané-Farré, Michel Debarbouille, Magdalena M van der Kooi-Pol, Cyprien Guérin, Sandra Dérozier, Aurelia Hiron, Hanne Jarmer, Aurélie Leduc, Stephan Michalik, Ewoud Reilman, Marc Schaffer, Frank Schmidt, Philippe Bessières, Philippe Noirot, Michael Hecker, Tarek Msadek, Uwe Völker, Jan Maarten van Dijl
Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions.
PLoS Genet: 2016, 12(4);e1005962
[PubMed:27035918] [WorldCat.org] [DOI] (I e)
