Jump to navigation
Jump to search
UniProt: 10-MAY-2017
⊟Summary[edit | edit source]
- organism: Staphylococcus aureus NCTC8325
- locus tag: SAOUHSC_01139/SAOUHSC_01141
- pan locus tag?: SAUPAN003446000
- symbol: bshC
- pan gene symbol?: bshC
- synonym:
- product: Putative cysteine ligase BshC
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
⊟Accession numbers[edit | edit source]
- Gene ID:
- RefSeq:
- BioCyc: G1I0R-1070 BioCyc
- MicrobesOnline: 1289595 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421
481
541
601
661
721
781
841
901
961
1021
1081
1141
1201
1261
1321
1381
1441
1501
1561
1621ATGGACTGTAAAGTAGTTAGTTTAAATGAAAAAGATCAGTTTATACCAAAAATAAAGAGC
AGTGACCCTGTAATAACAGGATTATTTCAATATGATGCAGCTCAACAAACTAGTTTTGAA
AAAAGGATGTCTAAAGAAAATAATGGAAGAGAAGCGGCATTAGCGAATGTTATTCGTGAA
TATATGAGTGATTTAAAGCTTTCAAGCTTTCAAGTGAACAAGAATTAAACATACAACATT
TAGCTAATGGTTCAAAAGTTGTGATTGGTGGACAACAAGCAGGGCTTTTCGGGGGACCAT
TGTATACATTCCATAAAATATTTTCAATCATTACTTTATCTAAGGAATTAACGGATACAC
ATAAGCAACAAGTAGTACCAGTTTTTTGGATTGCAGGAGAAGATCATGATTTCGATGAAG
TGAATCATACATTTGTTTATAACGAAAATCATGGGTCGCTGCATAAGGTTAAATATCATA
CAATGGAGATGCCAGAGACGACTGTCTCTAGATATTATCCTGATAAGGCTGAGTTGAAAC
AAACTTTAAAAACGATGTTCATTCATATGAAAGAAACTGTTCATACACAAGGTCTACTGG
AGATTTGTGACAGAATTATTGACCAATATGACTCGTGGACTGATATGTTTAAAGCACTAC
TGCATGAAACATTTAAAGCATATGGCGTTCTATTTATAGATGCGCAGTTTGAGCCGTTAA
GAAAAATGGAAGCGCCTATGTTTAAAAAGATTTTGAAAAAACATCAGTTGCTTGATGATG
CTTTTAGAGCAACACAACAACGTACTCAAAATCAAGGCTTGAATGCGATGATACAAACAG
ATACAAATGTTCATTTATTCTTACATGATGAAAATATGCGTCAATTAGTTTCGTATGATG
GTAAGCATTTTAAATTAAATAAAACAGATAAGACATATATAAAGGAAGAAATTATAAATA
TTGCGGAAAATCAACCTGAATTATTTTCTAATAATGTAGTGACAAGACCATTAATGGAAG
AATGGTTATTTAACACGGTGGCATTTGTTGGAGGACCGAGTGAAATTAAGTACTGGGCTG
AACTAAAAGATGTATTTGAACTATTTGATGTTGAAATGCCTATCGTGATGCCAAGGCTTA
GAATTACTTATTTAAATGACCGTATAGAAAAATTACTTTCGAAATACAATATTCCATTAG
AAAAAGTGTTAGTCGATGGTGTTGAAGGAGAAAGAAGTAAGTTTATTAGAGAACAAGCAT
CACATCAATTTATTGAAAAGGTAGAAGGTATGATTGAACAACAGCGTCGTCTAAACAAAG
ACTTATTAGATGAAGTGGCGGGGAATCAAAATAATATTAACCTTGTGAATAAAAATAATG
AAATTCATATACAACAGTATGATTATTTGTTAAAACGTTATCTTTTAAACATTGAAAGAG
AAAACGACATCAGTATGAAGCAATTTAGAGAAATTCAAGAAACACTCCATCCAATGGGAG
GATTACAAGAAAGAATATGGAATCCACTTCAAATTTTGAATGATTTTGGGACAGATGTGT
TCAAGCCCTCCACCTATCCACCACTTTCTTACACTTTTGATCATATTATTATAAAACCTT
AA60
120
180
240
300
360
420
480
540
600
660
720
780
840
900
960
1020
1080
1140
1200
1260
1320
1380
1440
1500
1560
1620
1622
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SAOUHSC_01139/SAOUHSC_01141
- symbol: BshC
- description: Putative cysteine ligase BshC
- length:
- theoretical pI:
- theoretical MW:
- GRAVY:
⊟Function[edit | edit source]
- reaction: EC 6.-.-.-? ExPASy
- TIGRFAM:
- TheSEED: data available for COL, N315, Newman, USA300_FPR3757
- PFAM:
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors:
- effectors:
⊟Localization[edit | edit source]
- PSORTb:
- DeepLocPro:
- LocateP:
- SignalP:
- predicted transmembrane helices (TMHMM):
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
⊟Experimental data[edit | edit source]
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
- predicted SigA promoter [2] : S470 > SAOUHSC_01135 > SAOUHSC_01136 > SAOUHSC_01137 > bshC
⊟Regulation[edit | edit source]
- regulator:
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: [2]
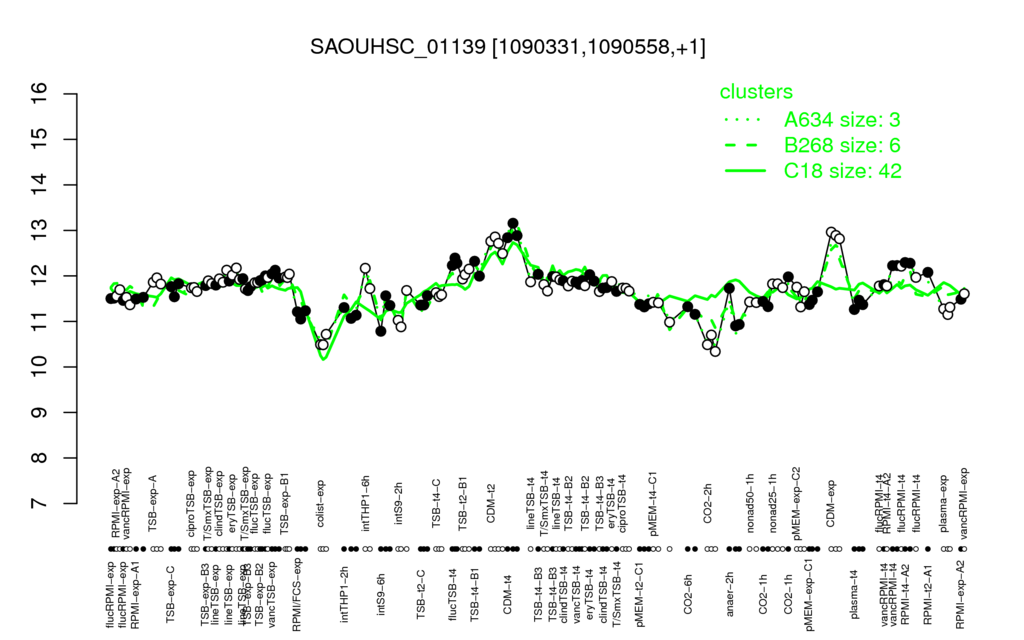 Multi-gene expression profiles
Multi-gene expression profiles
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
- Aureolib: no data available
⊟Protein stability[edit | edit source]
- half-life: no data available
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You can add further information about the gene and protein here. [edit]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ Dierk-Christoph Pöther, Philipp Gierok, Manuela Harms, Jörg Mostertz, Falko Hochgräfe, Haike Antelmann, Chris J Hamilton, Ilya Borovok, Michael Lalk, Yair Aharonowitz, Michael Hecker
Distribution and infection-related functions of bacillithiol in Staphylococcus aureus.
Int J Med Microbiol: 2013, 303(3);114-23
[PubMed:23517692] [WorldCat.org] [DOI] (I p) - ↑ 2.0 2.1 2.2 Ulrike Mäder, Pierre Nicolas, Maren Depke, Jan Pané-Farré, Michel Debarbouille, Magdalena M van der Kooi-Pol, Cyprien Guérin, Sandra Dérozier, Aurelia Hiron, Hanne Jarmer, Aurélie Leduc, Stephan Michalik, Ewoud Reilman, Marc Schaffer, Frank Schmidt, Philippe Bessières, Philippe Noirot, Michael Hecker, Tarek Msadek, Uwe Völker, Jan Maarten van Dijl
Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions.
PLoS Genet: 2016, 12(4);e1005962
[PubMed:27035918] [WorldCat.org] [DOI] (I e)
⊟Relevant publications[edit | edit source]
Dierk-Christoph Pöther, Philipp Gierok, Manuela Harms, Jörg Mostertz, Falko Hochgräfe, Haike Antelmann, Chris J Hamilton, Ilya Borovok, Michael Lalk, Yair Aharonowitz, Michael Hecker
Distribution and infection-related functions of bacillithiol in Staphylococcus aureus.
Int J Med Microbiol: 2013, 303(3);114-23
[PubMed:23517692] [WorldCat.org] [DOI] (I p)
