Jump to navigation
Jump to search
NCBI: 03-AUG-2016
⊟Summary[edit | edit source]
- organism: Staphylococcus aureus NCTC8325
- locus tag: SAOUHSC_01364
- pan locus tag?: SAUPAN003758000
- symbol: SAOUHSC_01364
- pan gene symbol?: tyrA
- synonym:
- product: prephenate dehydrogenase
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
- type: CDS
- locus tag: SAOUHSC_01364
- symbol: SAOUHSC_01364
- product: prephenate dehydrogenase
- replicon: chromosome
- strand: -
- coordinates: 1307658..1308749
- length: 1092
- essential: no DEG other strains
⊟Accession numbers[edit | edit source]
- Gene ID: 3920069 NCBI
- RefSeq: YP_499891 NCBI
- BioCyc: G1I0R-1274 BioCyc
- MicrobesOnline: 1289805 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421
481
541
601
661
721
781
841
901
961
1021
1081ATGACAACAGTTTTATTTGTTGGGCTTGGATTAATTGGTGGAAGTCTTGCTAGCAATATA
AAATACCATAACCCTAATACTAATATTATTGCATACGATGCAGATATTTCTCAGTTAGAT
AAAGCTAAATCAATCGGCATTATTAATGAAAAATGTTTAAATTATAGTGAAGCTATTAAA
AAAGCCGATGTAATTATTTATGCAACACCTGTTGCTATCACAAATAAATATCTTAGCGAG
CTTATAGATATGCCAACTAAACCTGGTGTTATTGTTTCTGATACTGGTAGTACTAAAGCA
ATGATACAGCAACACGAATGCAATTTATTAAAGCATAATATTCATTTAGTCAGTGGTCAT
CCAATGGCTGGTAGTCATAAATCTGGTGTACTAAATGCTAAAAAGCACTTATTTGAAAAC
GCTTATTATATTTTAGTCTACAATGAGCCAAGAAATGAGCAAGCAGCAAACACGTTAAAA
GAACTGTTATCACCTACTCTTGCTAAATTTATTGTAACTACTGCTGAAGAACACGACTAC
GTAACAAGCGTCGTAAGTCATTTACCTCATATCGTTGCATCTAGTTTAGTTCATGTTAGT
CAAAAGAACGGTCAAGAACATCATTTAGTTAATAAACTTGCAGCTGGTGGTTTTCGTGAT
ATCACTCGTATAGCTAGTAGTAATGCACAAATGTGGAAAGATATCACCTTGAGTAATAAA
ACGTATATTTTAGAAATGATTCGACAGCTAAAAAGTCAGTTTCAAGATTTAGAAAGACTA
ATTGAAAGCAATGATTCTGAAAAATTGTTATCATTTTTTGCCCAAGCTAAATCGTATCGC
GACGCACTACCCGCTAAACAACTAGGTGGACTAAATACTGCGTATGATCTATATGTAGAC
ATTCCGGATGAATCAGGTATGATAAGTAAAGTGACTTATATCCTGAGTTTACATAACATA
TCTATAAGCAACTTAAGAATCTTAGAAGTACGCGAAGATATATACGGTGCTTTAAAAATT
AGTTTCAAAAACCCTACTGACCGAGAACGCGGTATGCAAGCATTGAGTGATTTTGATTGT
TATATCCAATAA60
120
180
240
300
360
420
480
540
600
660
720
780
840
900
960
1020
1080
1092
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SAOUHSC_01364
- symbol: SAOUHSC_01364
- description: prephenate dehydrogenase
- length: 363
- theoretical pI: 7.22968
- theoretical MW: 40407
- GRAVY: -0.153168
⊟Function[edit | edit source]
- reaction: EC 1.3.1.12? ExPASyPrephenate dehydrogenase Prephenate + NAD+ = 4-hydroxyphenylpyruvate + CO2 + NADH
- TIGRFAM: Amino acid biosynthesis Serine family phosphoglycerate dehydrogenase (TIGR01327; EC 1.1.1.95; HMM-score: 26.7)and 5 moreEnergy metabolism Pentose phosphate pathway 6-phosphogluconate dehydrogenase (decarboxylating) (TIGR00872; EC 1.1.1.44; HMM-score: 20.6)Amino acid biosynthesis Other pyrrolysine biosynthesis protein PylD (TIGR03911; HMM-score: 17.9)Amino acid biosynthesis Glutamate family pyrroline-5-carboxylate reductase (TIGR00112; EC 1.5.1.2; HMM-score: 15.4)Energy metabolism Amino acids and amines 3-hydroxyisobutyrate dehydrogenase (TIGR01692; EC 1.1.1.31; HMM-score: 14.1)Amino acid biosynthesis Pyruvate family ketol-acid reductoisomerase (TIGR00465; EC 1.1.1.86; HMM-score: 11.4)
- TheSEED :
- Prephenate dehydrogenase (EC 1.3.1.12)
Amino Acids and Derivatives Aromatic amino acids and derivatives Chorismate Synthesis Prephenate dehydrogenase (EC 1.3.1.12)and 1 more - PFAM: NADP_Rossmann (CL0063) PDH_N; Prephenate dehydrogenase, nucleotide-binding domain (PF02153; HMM-score: 108.1)6PGD_C (CL0106) PDH_C; Prephenate dehydrogenase, dimerization domain (PF20463; HMM-score: 104.5)and 12 moreNADP_Rossmann (CL0063) F420_oxidored; NADP oxidoreductase coenzyme F420-dependent (PF03807; HMM-score: 31.2)NAD_binding_2; NAD binding domain of 6-phosphogluconate dehydrogenase (PF03446; HMM-score: 23.3)ACT (CL0070) ACT; ACT domain (PF01842; HMM-score: 20.1)NADP_Rossmann (CL0063) 2-Hacid_dh_C; D-isomer specific 2-hydroxyacid dehydrogenase, NAD binding domain (PF02826; HMM-score: 19.5)KARI_N; Acetohydroxy acid isomeroreductase, NADPH-binding domain (PF07991; HMM-score: 15.7)no clan defined P4Ha_N; Prolyl 4-Hydroxylase alpha-subunit, N-terminal region (PF08336; HMM-score: 15.1)NADP_Rossmann (CL0063) GFO_IDH_MocA; Oxidoreductase family, NAD-binding Rossmann fold (PF01408; HMM-score: 15)ACT (CL0070) ACT_AHAS_ss; AHAS small subunit-like ACT domain (PF22629; HMM-score: 14.9)no clan defined SbcD_C; Type 5 capsule protein repressor C-terminal domain (PF12320; HMM-score: 12.9)NADP_Rossmann (CL0063) TrkA_N; TrkA-N domain (PF02254; HMM-score: 12.7)Sacchrp_dh_NADP; Saccharopine dehydrogenase NADP binding domain (PF03435; HMM-score: 12.7)Spectrin (CL0743) Spectrin_2; Spectrin like domain (PF18373; HMM-score: 11.8)
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors:
- effectors:
⊟Localization[edit | edit source]
- PSORTb: Cytoplasmic
- Cytoplasmic Score: 7.5
- Cytoplasmic Membrane Score: 1.15
- Cellwall Score: 0.62
- Extracellular Score: 0.73
- Internal Helices: 0
- DeepLocPro: Cytoplasmic
- Cytoplasmic Score: 0.9291
- Cytoplasmic Membrane Score: 0.0531
- Cell wall & surface Score: 0.001
- Extracellular Score: 0.0168
- LocateP: Intracellular
- Prediction by SwissProt Classification: Cytoplasmic
- Pathway Prediction: No pathway
- Intracellular possibility: 0.83
- Signal peptide possibility: -0.5
- N-terminally Anchored Score: -2
- Predicted Cleavage Site: No CleavageSite
- SignalP: no predicted signal peptide
- SP(Sec/SPI): 0.37858
- TAT(Tat/SPI): 0.003113
- LIPO(Sec/SPII): 0.130738
- predicted transmembrane helices (TMHMM): 0
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MTTVLFVGLGLIGGSLASNIKYHNPNTNIIAYDADISQLDKAKSIGIINEKCLNYSEAIKKADVIIYATPVAITNKYLSELIDMPTKPGVIVSDTGSTKAMIQQHECNLLKHNIHLVSGHPMAGSHKSGVLNAKKHLFENAYYILVYNEPRNEQAANTLKELLSPTLAKFIVTTAEEHDYVTSVVSHLPHIVASSLVHVSQKNGQEHHLVNKLAAGGFRDITRIASSNAQMWKDITLSNKTYILEMIRQLKSQFQDLERLIESNDSEKLLSFFAQAKSYRDALPAKQLGGLNTAYDLYVDIPDESGMISKVTYILSLHNISISNLRILEVREDIYGALKISFKNPTDRERGMQALSDFDCYIQ
⊟Experimental data[edit | edit source]
- experimentally validated: PeptideAtlas [1] [2]
- protein localization: data available for COL
- quantitative data / protein copy number per cell:
- interaction partners:
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
- MicrobesOnline: no polycistronic organisation predicted
⊟Regulation[edit | edit source]
- regulator: CodY* (repression) regulon
CodY* (TF) important in Amino acid metabolism; RegPrecise
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: [3]
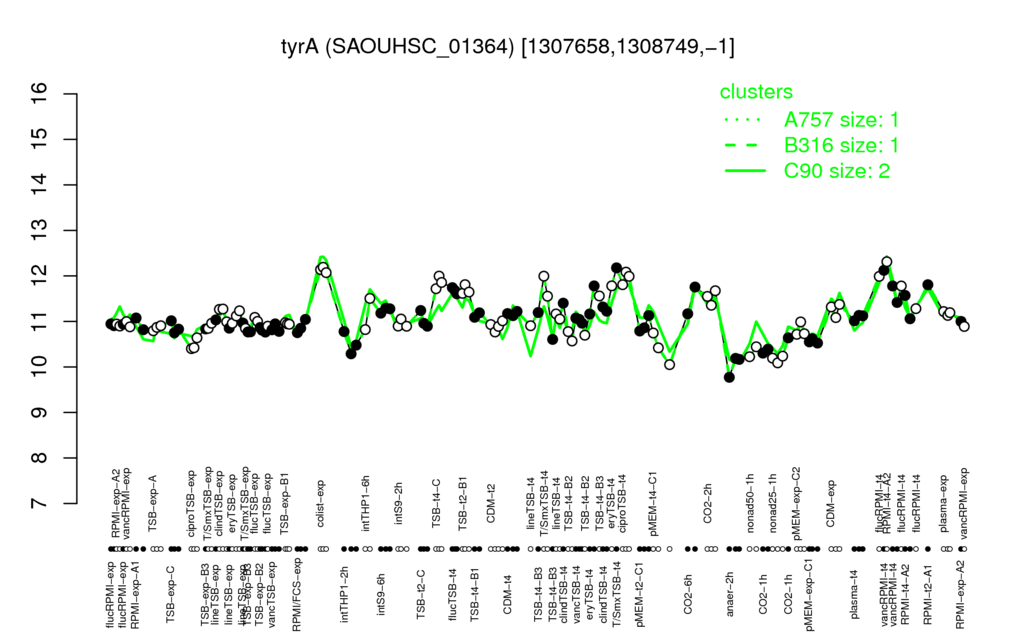 Multi-gene expression profiles
Multi-gene expression profiles
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
- Aureolib: no data available
⊟Protein stability[edit | edit source]
- half-life: no data available
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You can add further information about the gene and protein here. [edit]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ Maren Depke, Stephan Michalik, Alexander Rabe, Kristin Surmann, Lars Brinkmann, Nico Jehmlich, Jörg Bernhardt, Michael Hecker, Bernd Wollscheid, Zhi Sun, Robert L Moritz, Uwe Völker, Frank Schmidt
A peptide resource for the analysis of Staphylococcus aureus in host-pathogen interaction studies.
Proteomics: 2015, 15(21);3648-61
[PubMed:26224020] [WorldCat.org] [DOI] (I p) - ↑ Stephan Michalik, Maren Depke, Annette Murr, Manuela Gesell Salazar, Ulrike Kusebauch, Zhi Sun, Tanja C Meyer, Kristin Surmann, Henrike Pförtner, Petra Hildebrandt, Stefan Weiss, Laura Marcela Palma Medina, Melanie Gutjahr, Elke Hammer, Dörte Becher, Thomas Pribyl, Sven Hammerschmidt, Eric W Deutsch, Samuel L Bader, Michael Hecker, Robert L Moritz, Ulrike Mäder, Uwe Völker, Frank Schmidt
A global Staphylococcus aureus proteome resource applied to the in vivo characterization of host-pathogen interactions.
Sci Rep: 2017, 7(1);9718
[PubMed:28887440] [WorldCat.org] [DOI] (I e) - ↑ Ulrike Mäder, Pierre Nicolas, Maren Depke, Jan Pané-Farré, Michel Debarbouille, Magdalena M van der Kooi-Pol, Cyprien Guérin, Sandra Dérozier, Aurelia Hiron, Hanne Jarmer, Aurélie Leduc, Stephan Michalik, Ewoud Reilman, Marc Schaffer, Frank Schmidt, Philippe Bessières, Philippe Noirot, Michael Hecker, Tarek Msadek, Uwe Völker, Jan Maarten van Dijl
Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions.
PLoS Genet: 2016, 12(4);e1005962
[PubMed:27035918] [WorldCat.org] [DOI] (I e)
