Jump to navigation
Jump to search
NCBI: 03-AUG-2016
⊟Summary[edit | edit source]
- organism: Staphylococcus aureus NCTC8325
- locus tag: SAOUHSC_02333
- pan locus tag?: SAUPAN005381000
- symbol: SAOUHSC_02333
- pan gene symbol?: sceD
- synonym:
- product: transglycosylase SceD
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
- type: CDS
- locus tag: SAOUHSC_02333
- symbol: SAOUHSC_02333
- product: transglycosylase SceD
- replicon: chromosome
- strand: -
- coordinates: 2162622..2163317
- length: 696
- essential: no DEG other strains
⊟Accession numbers[edit | edit source]
- Gene ID: 3920958 NCBI
- RefSeq: YP_500812 NCBI
- BioCyc: G1I0R-2205 BioCyc
- MicrobesOnline: 1290773 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421
481
541
601
661ATGAAGAAAACATTACTCGCATCATCATTAGCAGTAGGTTTAGGAATCGTAGCAGGAAAT
GCAGGTCACGAAGCCCATGCAAGTGAAGCGGACTTAAATAAAGCATCTTTAGCGCAAATG
GCGCAATCAAATGATCAAACATTAAATCAAAAACCAATTGAAGCTGGGGCTTATAATTAT
ACATTTGACTATGAAGGGTTTACTTATCACTTTGAATCAGATGGTACACACTTTGCTTGG
AATTACCATGCAACAGGTACTAATGGAGCAGACATGAGTGCACAAGCACCTGCAACTAAT
AATGTTGCACCATCAGCTGTTCAAGCTAATCAAGTACAATCACAAGAAGTTGAAGCACCA
CAAAATGCTCAAACTCAACAACCACAAGCATCAACATCAAACAATTCACAAGTTACTGCA
ACACCAACTGAATCAAAATCATCAGAAGGTTCATCAGTAAATGTGAATGCTCATCTAAAA
CAAATTGCTCAACGTGAATCAGGTGGCAATATTCATGCTGTAAATCCAACATCAGGTGCA
GCTGGTAAGTATCAATTCTTACAATCAACTTGGGATTCAGTAGCACCTGCTAAATATAAA
GGTGTATCACCAGCAAATGCTCCTGAAAGTGTTCAAGATGCCGCAGCAGTAAAATTATAT
AACACTGGTGGCGCTGGACATTGGGTTACTGCATAA60
120
180
240
300
360
420
480
540
600
660
696
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SAOUHSC_02333
- symbol: SAOUHSC_02333
- description: transglycosylase SceD
- length: 231
- theoretical pI: 5.79248
- theoretical MW: 24065.9
- GRAVY: -0.546753
⊟Function[edit | edit source]
- reaction: EC 3.2.-.-? ExPASy
- TIGRFAM:
- TheSEED :
- SceD-like transglycosylase, biomarker for vancomycin-intermediate strains
- PFAM: Lysozyme (CL0037) Transglycosylas; Transglycosylase-like domain (PF06737; HMM-score: 89.8)and 4 moreSLT; Transglycosylase SLT domain (PF01464; HMM-score: 23.6)no clan defined SIDD_N; Nonribosomal peptide synthetase sidD N-terminal (PF24895; HMM-score: 13.4)FtsA_C; Cell division protein FtsA, C-terminal (PF11983; HMM-score: 7.8)Lin-8; Ras-mediated vulval-induction antagonist (PF03353; HMM-score: 6.5)
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors:
- effectors:
⊟Localization[edit | edit source]
- PSORTb: Extracellular
- Cytoplasmic Score: 0
- Cytoplasmic Membrane Score: 0
- Cellwall Score: 0.02
- Extracellular Score: 9.98
- Internal Helices: 0
- DeepLocPro: Extracellular
- Cytoplasmic Score: 0.0001
- Cytoplasmic Membrane Score: 0.0004
- Cell wall & surface Score: 0.0747
- Extracellular Score: 0.9248
- LocateP: Secretory(released) (with CS)
- Prediction by SwissProt Classification: Extracellular
- Pathway Prediction: Sec-(SPI)
- Intracellular possibility: -0.17
- Signal peptide possibility: 1
- N-terminally Anchored Score: -2
- Predicted Cleavage Site: HEAHASEA
- SignalP: Signal peptide SP(Sec/SPI) length 27 aa
- SP(Sec/SPI): 0.986312
- TAT(Tat/SPI): 0.003284
- LIPO(Sec/SPII): 0.008779
- Cleavage Site: CS pos: 27-28. AHA-SE. Pr: 0.9175
- predicted transmembrane helices (TMHMM): 0
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MKKTLLASSLAVGLGIVAGNAGHEAHASEADLNKASLAQMAQSNDQTLNQKPIEAGAYNYTFDYEGFTYHFESDGTHFAWNYHATGTNGADMSAQAPATNNVAPSAVQANQVQSQEVEAPQNAQTQQPQASTSNNSQVTATPTESKSSEGSSVNVNAHLKQIAQRESGGNIHAVNPTSGAAGKYQFLQSTWDSVAPAKYKGVSPANAPESVQDAAAVKLYNTGGAGHWVTA
⊟Experimental data[edit | edit source]
- experimentally validated: PeptideAtlas [1] [2]
- protein localization: data available for COL
- quantitative data / protein copy number per cell:
- interaction partners:
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
- MicrobesOnline: no polycistronic organisation predicted
⊟Regulation[edit | edit source]
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: [4]
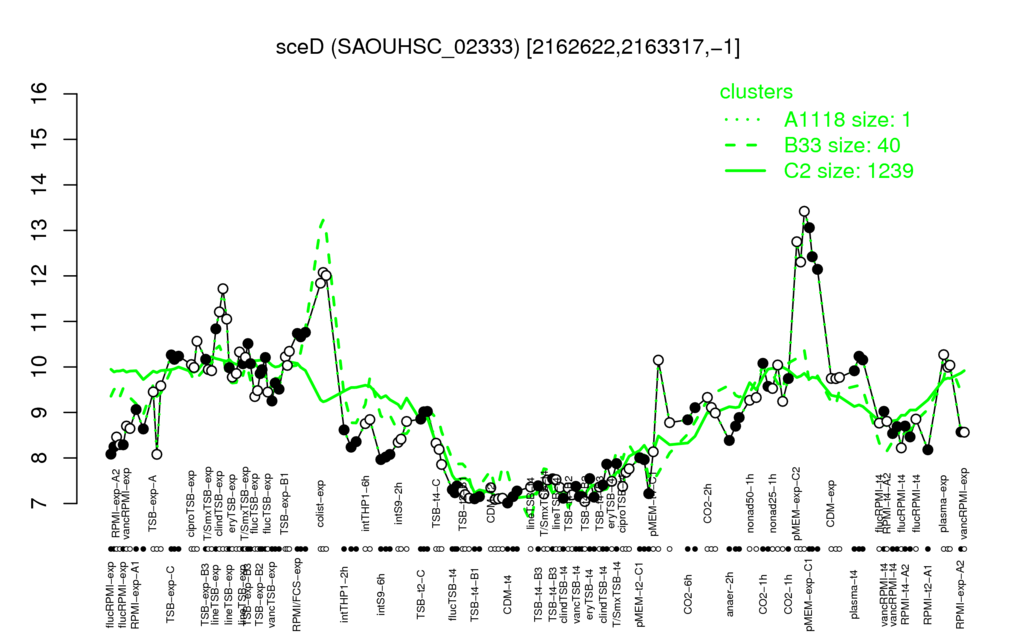 Multi-gene expression profiles
Multi-gene expression profiles
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
- Aureolib: no data available
⊟Protein stability[edit | edit source]
- half-life: no data available
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You can add further information about the gene and protein here. [edit]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ Maren Depke, Stephan Michalik, Alexander Rabe, Kristin Surmann, Lars Brinkmann, Nico Jehmlich, Jörg Bernhardt, Michael Hecker, Bernd Wollscheid, Zhi Sun, Robert L Moritz, Uwe Völker, Frank Schmidt
A peptide resource for the analysis of Staphylococcus aureus in host-pathogen interaction studies.
Proteomics: 2015, 15(21);3648-61
[PubMed:26224020] [WorldCat.org] [DOI] (I p) - ↑ Stephan Michalik, Maren Depke, Annette Murr, Manuela Gesell Salazar, Ulrike Kusebauch, Zhi Sun, Tanja C Meyer, Kristin Surmann, Henrike Pförtner, Petra Hildebrandt, Stefan Weiss, Laura Marcela Palma Medina, Melanie Gutjahr, Elke Hammer, Dörte Becher, Thomas Pribyl, Sven Hammerschmidt, Eric W Deutsch, Samuel L Bader, Michael Hecker, Robert L Moritz, Ulrike Mäder, Uwe Völker, Frank Schmidt
A global Staphylococcus aureus proteome resource applied to the in vivo characterization of host-pathogen interactions.
Sci Rep: 2017, 7(1);9718
[PubMed:28887440] [WorldCat.org] [DOI] (I e) - ↑ Sarah Dubrac, Ivo Gomperts Boneca, Olivier Poupel, Tarek Msadek
New insights into the WalK/WalR (YycG/YycF) essential signal transduction pathway reveal a major role in controlling cell wall metabolism and biofilm formation in Staphylococcus aureus.
J Bacteriol: 2007, 189(22);8257-69
[PubMed:17827301] [WorldCat.org] [DOI] (I p) - ↑ Ulrike Mäder, Pierre Nicolas, Maren Depke, Jan Pané-Farré, Michel Debarbouille, Magdalena M van der Kooi-Pol, Cyprien Guérin, Sandra Dérozier, Aurelia Hiron, Hanne Jarmer, Aurélie Leduc, Stephan Michalik, Ewoud Reilman, Marc Schaffer, Frank Schmidt, Philippe Bessières, Philippe Noirot, Michael Hecker, Tarek Msadek, Uwe Völker, Jan Maarten van Dijl
Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions.
PLoS Genet: 2016, 12(4);e1005962
[PubMed:27035918] [WorldCat.org] [DOI] (I e)
⊟Relevant publications[edit | edit source]
Melanie R Stapleton, Malcolm J Horsburgh, Emma J Hayhurst, Lynda Wright, Ing-Marie Jonsson, Andrej Tarkowski, John F Kokai-Kun, James J Mond, Simon J Foster
Characterization of IsaA and SceD, two putative lytic transglycosylases of Staphylococcus aureus.
J Bacteriol: 2007, 189(20);7316-25
[PubMed:17675373] [WorldCat.org] [DOI] (P p)
