⊟Summary[edit | edit source]
- organism: Staphylococcus aureus NCTC8325
- locus tag: SAOUHSC_02963
- pan locus tag?: SAUPAN006357000
- symbol: SAOUHSC_02963
- pan gene symbol?: clfB
- synonym:
- product: clumping factor B
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
- type: CDS
- locus tag: SAOUHSC_02963
- symbol: SAOUHSC_02963
- product: clumping factor B
- replicon: chromosome
- strand: -
- coordinates: 2722968..2725601
- length: 2634
- essential: no DEG other strains
⊟Accession numbers[edit | edit source]
- Gene ID: 3921665 NCBI
- RefSeq: YP_501414 NCBI
- BioCyc: G1I0R-2787 BioCyc
- MicrobesOnline: 1291385 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421
481
541
601
661
721
781
841
901
961
1021
1081
1141
1201
1261
1321
1381
1441
1501
1561
1621
1681
1741
1801
1861
1921
1981
2041
2101
2161
2221
2281
2341
2401
2461
2521
2581TTGAAAAAAAGAATTGATTATTTGTCGAATAAGCAGAATAAGTATTCGATTAGACGTTTT
ACAGTAGGTACCACATCAGTAATAGTAGGGGCAACTATACTATTTGGGATAGGCAATCAT
CAAGCACAAGCTTCAGAACAATCGAACGATACAACGCAATCTTCGAAAAATAATGCAAGT
GCAGATTCCGAAAAAAACAATATGATAGAAACACCTCAATTAAATACAACGGCTAATGAT
ACATCTGATATTAGTGCAAACACAAACAGTGCGAATGTAGATAGCACAACAAAACCAATG
TCTACACAAACGAGCAATACCACTACAACAGAGCCAGCTTCAACAAATGAAACACCTCAA
CCGACGGCAATTAAAAATCAAGCAACTGCTGCAAAAATGCAAGATCAAACTGTTCCTCAA
GAAGCAAATTCTCAAGTAGATAATAAAACAACGAATGATGCTAATAGCATAGCAACAAAC
AGTGAGCTTAAAAATTCTCAAACATTAGATTTACCACAATCATCACCACAAACGATTTCC
AATGCGCAAGGAACTAGTAAACCAAGTGTTAGAACGAGAGCTGTACGTAGTTTAGCTGTT
GCTGAACCGGTAGTAAATGCTGCTGATGCTAAAGGTACAAATGTAAATGATAAAGTTACG
GCAAGTAATTTCAAGTTAGAAAAGACTACATTTGACCCTAATCAAAGTGGTAACACATTT
ATGGCGGCAAATTTTACAGTGACAGATAAAGTGAAATCAGGGGATTATTTTACAGCGAAG
TTACCAGATAGTTTAACTGGTAATGGAGACGTGGATTATTCTAATTCAAATAATACGATG
CCAATTGCAGACATTAAAAGTACGAATGGCGATGTTGTAGCTAAAGCAACATATGATATC
TTGACTAAGACGTATACATTTGTCTTTACAGATTATGTAAATAATAAAGAAAATATTAAC
GGACAATTTTCATTACCTTTATTTACAGACCGAGCAAAGGCACCTAAATCAGGAACATAT
GATGCGAATATTAATATTGCGGATGAAATGTTTAATAATAAAATTACTTATAACTATAGT
TCGCCAATTGCAGGAATTGATAAACCAAATGGCGCGAACATTTCTTCTCAAATTATTGGT
GTAGATACAGCTTCAGGTCAAAACACATACAAGCAAACAGTATTTGTTAACCCTAAGCAA
CGAGTTTTAGGTAATACGTGGGTGTATATTAAAGGCTACCAAGATAAAATCGAAGAAAGT
AGCGGTAAAGTAAGTGCTACAGATACAAAACTGAGAATTTTTGAAGTGAATGATACATCT
AAATTATCAGATAGCTACTATGCAGATCCAAATGACTCTAACCTTAAAGAAGTAACAGAC
CAATTTAAAAATAGAATCTATTATGAGCATCCAAATGTAGCTAGTATTAAATTTGGTGAT
ATTACTAAAACATATGTAGTATTAGTAGAAGGGCATTACGACAATACAGGTAAGAACTTA
AAAACTCAGGTTATTCAAGAAAATGTTGATCCTGTAACAAATAGAGACTACAGTATTTTC
GGTTGGAATAATGAGAATGTTGTACGTTATGGTGGTGGAAGTGCTGATGGTGATTCAGCA
GTAAATCCGAAAGACCCAACTCCAGGGCCGCCGGTTGACCCAGAACCAAGTCCAGACCCA
GAACCAGAACCAACGCCAGATCCAGAACCAAGTCCAGACCCAGAACCGGAACCAAGCCCA
GACCCGGATCCGGATTCGGATTCAGACAGTGACTCAGGCTCAGACAGCGACTCAGGTTCA
GATAGCGACTCAGAATCAGATAGCGATTCGGATTCAGACAGTGATTCAGATTCAGACAGC
GACTCAGAATCAGATAGCGACTCAGAATCAGATAGTGAGTCAGATTCAGACAGTGACTCG
GACTCAGACAGTGATTCAGACTCAGATAGCGATTCAGACTCAGATAGCGATTCAGACTCA
GACAGCGATTCAGATTCAGACAGCGACTCAGATTCAGACAGCGACTCAGACTCAGATAGC
GACTCAGACTCAGACAGCGACTCAGATTCAGATAGCGATTCAGACTCAGACAGCGACTCA
GACTCAGACAGCGACTCAGACTCAGATAGCGACTCAGATTCAGATAGCGATTCAGACTCA
GACAGCGACTCAGATTCAGATAGCGATTCGGACTCAGACAGCGATTCAGATTCAGACAGC
GACTCAGACTCGGATAGCGATTCAGATTCAGATAGCGATTCGGATTCAGACAGTGATTCA
GATTCAGACAGCGACTCAGACTCGGATAGCGACTCAGACTCAGACAGCGATTCAGACTCA
GATAGCGACTCAGACTCGGATAGCGACTCGGATTCAGATAGCGACTCAGACTCAGATAGT
GACTCCGATTCAAGAGTTACACCACCAAATAATGAACAGAAAGCACCATCAAATCCTAAA
GGTGAAGTAAACCATTCTAATAAGGTATCAAAACAACACAAAACTGATGCTTTACCAGAA
ACAGGAGATAAGAGCGAAAACACAAATGCAACTTTATTTGGTGCAATGATGGCATTATTA
GGATCATTACTATTGTTTAGAAAACGCAAGCAAGATCATAAAGAAAAAGCGTAA60
120
180
240
300
360
420
480
540
600
660
720
780
840
900
960
1020
1080
1140
1200
1260
1320
1380
1440
1500
1560
1620
1680
1740
1800
1860
1920
1980
2040
2100
2160
2220
2280
2340
2400
2460
2520
2580
2634
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SAOUHSC_02963
- symbol: SAOUHSC_02963
- description: clumping factor B
- length: 877
- theoretical pI: 3.73123
- theoretical MW: 93593.9
- GRAVY: -1.16956
⊟Function[edit | edit source]
- TIGRFAM: gram-positive signal peptide, YSIRK family (TIGR01168; HMM-score: 52)and 1 moreCell envelope Other LPXTG cell wall anchor domain (TIGR01167; HMM-score: 28.7)
- TheSEED :
- Clumping factor ClfB, fibrinogen binding protein
- PFAM: Adhesin (CL0204) SdrG_C_C; C-terminus of bacterial fibrinogen-binding adhesin (PF10425; HMM-score: 148.8)and 4 moreE-set (CL0159) Big_8; Bacterial Ig domain (PF17961; HMM-score: 87.3)no clan defined YSIRK_signal; YSIRK type signal peptide (PF04650; HMM-score: 44.1)Gram_pos_anchor; LPXTG cell wall anchor motif (PF00746; HMM-score: 40.2)GBD (CL0202) Androglobin_IV; Androglobin, domain IV (PF22069; HMM-score: 15.2)
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors:
- effectors:
⊟Localization[edit | edit source]
- PSORTb: Cellwall
- Cytoplasmic Score: 0
- Cytoplasmic Membrane Score: 0
- Cellwall Score: 10
- Extracellular Score: 0
- Internal Helices: 2
- DeepLocPro: Cell wall & surface
- Cytoplasmic Score: 0.0001
- Cytoplasmic Membrane Score: 0.0046
- Cell wall & surface Score: 0.8889
- Extracellular Score: 0.1064
- LocateP: LPxTG Cell-wall anchored
- Prediction by SwissProt Classification: Cell Wall
- Pathway Prediction: Sec-(SPI)
- Intracellular possibility: 0.17
- Signal peptide possibility: 0.5
- N-terminally Anchored Score: 2
- Predicted Cleavage Site: found LPxTG motif :LPETG
- SignalP: Signal peptide SP(Sec/SPI) length 44 aa
- SP(Sec/SPI): 0.899149
- TAT(Tat/SPI): 0.06812
- LIPO(Sec/SPII): 0.003061
- Cleavage Site: CS pos: 44-45. AQA-SE. Pr: 0.9420
- predicted transmembrane helices (TMHMM): 1
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MKKRIDYLSNKQNKYSIRRFTVGTTSVIVGATILFGIGNHQAQASEQSNDTTQSSKNNASADSEKNNMIETPQLNTTANDTSDISANTNSANVDSTTKPMSTQTSNTTTTEPASTNETPQPTAIKNQATAAKMQDQTVPQEANSQVDNKTTNDANSIATNSELKNSQTLDLPQSSPQTISNAQGTSKPSVRTRAVRSLAVAEPVVNAADAKGTNVNDKVTASNFKLEKTTFDPNQSGNTFMAANFTVTDKVKSGDYFTAKLPDSLTGNGDVDYSNSNNTMPIADIKSTNGDVVAKATYDILTKTYTFVFTDYVNNKENINGQFSLPLFTDRAKAPKSGTYDANINIADEMFNNKITYNYSSPIAGIDKPNGANISSQIIGVDTASGQNTYKQTVFVNPKQRVLGNTWVYIKGYQDKIEESSGKVSATDTKLRIFEVNDTSKLSDSYYADPNDSNLKEVTDQFKNRIYYEHPNVASIKFGDITKTYVVLVEGHYDNTGKNLKTQVIQENVDPVTNRDYSIFGWNNENVVRYGGGSADGDSAVNPKDPTPGPPVDPEPSPDPEPEPTPDPEPSPDPEPEPSPDPDPDSDSDSDSGSDSDSGSDSDSESDSDSDSDSDSDSDSDSESDSDSESDSESDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSRVTPPNNEQKAPSNPKGEVNHSNKVSKQHKTDALPETGDKSENTNATLFGAMMALLGSLLLFRKRKQDHKEKA
⊟Experimental data[edit | edit source]
- experimentally validated: PeptideAtlas [1] [2]
- protein localization: data available for COL
- quantitative data / protein copy number per cell:
- interaction partners:
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
- MicrobesOnline: no polycistronic organisation predicted
⊟Regulation[edit | edit source]
- regulator:
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: [3]
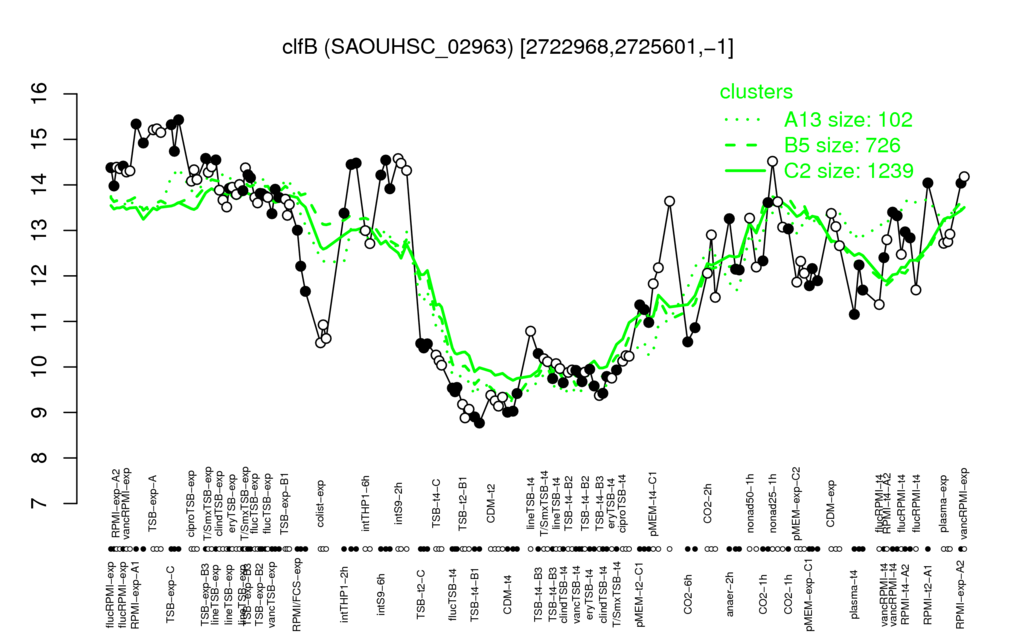 Multi-gene expression profiles
Multi-gene expression profiles
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
- Aureolib: no data available
⊟Protein stability[edit | edit source]
- half-life: no data available
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You can add further information about the gene and protein here. [edit]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ Maren Depke, Stephan Michalik, Alexander Rabe, Kristin Surmann, Lars Brinkmann, Nico Jehmlich, Jörg Bernhardt, Michael Hecker, Bernd Wollscheid, Zhi Sun, Robert L Moritz, Uwe Völker, Frank Schmidt
A peptide resource for the analysis of Staphylococcus aureus in host-pathogen interaction studies.
Proteomics: 2015, 15(21);3648-61
[PubMed:26224020] [WorldCat.org] [DOI] (I p) - ↑ Stephan Michalik, Maren Depke, Annette Murr, Manuela Gesell Salazar, Ulrike Kusebauch, Zhi Sun, Tanja C Meyer, Kristin Surmann, Henrike Pförtner, Petra Hildebrandt, Stefan Weiss, Laura Marcela Palma Medina, Melanie Gutjahr, Elke Hammer, Dörte Becher, Thomas Pribyl, Sven Hammerschmidt, Eric W Deutsch, Samuel L Bader, Michael Hecker, Robert L Moritz, Ulrike Mäder, Uwe Völker, Frank Schmidt
A global Staphylococcus aureus proteome resource applied to the in vivo characterization of host-pathogen interactions.
Sci Rep: 2017, 7(1);9718
[PubMed:28887440] [WorldCat.org] [DOI] (I e) - ↑ Ulrike Mäder, Pierre Nicolas, Maren Depke, Jan Pané-Farré, Michel Debarbouille, Magdalena M van der Kooi-Pol, Cyprien Guérin, Sandra Dérozier, Aurelia Hiron, Hanne Jarmer, Aurélie Leduc, Stephan Michalik, Ewoud Reilman, Marc Schaffer, Frank Schmidt, Philippe Bessières, Philippe Noirot, Michael Hecker, Tarek Msadek, Uwe Völker, Jan Maarten van Dijl
Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions.
PLoS Genet: 2016, 12(4);e1005962
[PubMed:27035918] [WorldCat.org] [DOI] (I e)
⊟Relevant publications[edit | edit source]
T J Foster, M Höök
Surface protein adhesins of Staphylococcus aureus.
Trends Microbiol: 1998, 6(12);484-8
[PubMed:10036727] [WorldCat.org] [DOI] (P p)F M McAleese, E J Walsh, M Sieprawska, J Potempa, T J Foster
Loss of clumping factor B fibrinogen binding activity by Staphylococcus aureus involves cessation of transcription, shedding and cleavage by metalloprotease.
J Biol Chem: 2001, 276(32);29969-78
[PubMed:11399757] [WorldCat.org] [DOI] (P p)Louise M O'Brien, Evelyn J Walsh, Ruth C Massey, Sharon J Peacock, Timothy J Foster
Staphylococcus aureus clumping factor B (ClfB) promotes adherence to human type I cytokeratin 10: implications for nasal colonization.
Cell Microbiol: 2002, 4(11);759-70
[PubMed:12427098] [WorldCat.org] [DOI] (P p)Evelyn J Walsh, Louise M O'Brien, Xiaowen Liang, Magnus Hook, Timothy J Foster
Clumping factor B, a fibrinogen-binding MSCRAMM (microbial surface components recognizing adhesive matrix molecules) adhesin of Staphylococcus aureus, also binds to the tail region of type I cytokeratin 10.
J Biol Chem: 2004, 279(49);50691-9
[PubMed:15385531] [WorldCat.org] [DOI] (P p)Heiman F L Wertheim, Evelyn Walsh, Roos Choudhurry, Damian C Melles, Hélène A M Boelens, Helen Miajlovic, Henri A Verbrugh, Timothy Foster, Alex van Belkum
Key role for clumping factor B in Staphylococcus aureus nasal colonization of humans.
PLoS Med: 2008, 5(1);e17
[PubMed:18198942] [WorldCat.org] [DOI] (I p)Ting Xue, Yibo You, Fei Shang, Baolin Sun
Rot and Agr system modulate fibrinogen-binding ability mainly by regulating clfB expression in Staphylococcus aureus NCTC8325.
Med Microbiol Immunol: 2012, 201(1);81-92
[PubMed:21701848] [WorldCat.org] [DOI] (I p)
