Jump to navigation
Jump to search
NCBI: 03-AUG-2016
⊟Summary[edit | edit source]
- organism: Staphylococcus aureus NCTC8325
- locus tag: SAOUHSC_01778
- pan locus tag?: SAUPAN004290000
- symbol: clpX
- pan gene symbol?: clpX
- synonym:
- product: ATP-dependent protease ATP-binding subunit ClpX
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
- type: CDS
- locus tag: SAOUHSC_01778
- symbol: clpX
- product: ATP-dependent protease ATP-binding subunit ClpX
- replicon: chromosome
- strand: -
- coordinates: 1677874..1679136
- length: 1263
- essential: no DEG other strains
⊟Accession numbers[edit | edit source]
- Gene ID: 3919696 NCBI
- RefSeq: YP_500283 NCBI
- BioCyc: G1I0R-1652 BioCyc
- MicrobesOnline: 1290197 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421
481
541
601
661
721
781
841
901
961
1021
1081
1141
1201
1261ATGTTTAAATTCAATGAAGATGAAGAAAATTTGAAATGCTCTTTCTGCGGAAAAGACCAA
GATCAAGTAAAAAAACTTGTAGCAGGAAGTGGTGTATATATTTGTAATGAGTGTATTGAA
TTATGCTCAGAAATCGTCGAAGAAGAATTAGCTCAAAACACTTCTGAAGCGATGACAGAA
TTACCTACTCCTAAAGAAATTATGGATCATTTAAACGAATATGTTATTGGTCAAGAAAAA
GCTAAAAAATCTTTAGCTGTAGCTGTTTATAACCACTATAAGCGTATTCAACAATTAGGA
CCAAAAGAAGATGATGTTGAATTACAAAAAAGTAACATTGCATTAATTGGGCCAACAGGT
AGTGGTAAAACATTATTAGCTCAAACCTTAGCCAAGACGTTGAATGTACCATTTGCAATT
GCAGATGCGACAAGTTTAACTGAAGCTGGTTATGTAGGCGATGATGTTGAAAATATCTTG
TTGAGATTAATTCAAGCAGCTGACTTTGACATTGATAAAGCCGAAAAAGGTATTATTTAT
GTAGATGAAATTGATAAAATTGCACGTAAATCTGAAAACACATCTATAACACGTGACGTT
TCAGGTGAAGGTGTTCAACAAGCATTGCTTAAAATCTTAGAAGGTACGACTGCAAGTGTT
CCGCCACAAGGTGGACGCAAACATCCAAACCAAGAAATGATTCAAATTGATACAACAAAT
ATCTTATTTATTCTTGGTGGTGCCTTTGATGGTATTGAAGAAGTGATTAAGCGCCGTCTT
GGTGAAAAAGTTATTGGTTTCTCAAGCAATGAAGCTGATAAATATGACGAACAAGCATTA
TTAGCACAAATTCGCCCAGAAGATTTGCAAGCCTATGGTTTGATTCCTGAATTTATCGGA
CGTGTGCCAATTGTAGCTAATTTAGAAACATTAGATGTAACTGCGTTGAAAAACATCTTA
ACGCAACCTAAAAATGCACTTGTGAAACAATATACTAAAATGCTGGAATTAGATGATGTG
GATTTAGAGTTCACTGAAGAAGCTTTATCAGCAATTAGTGAAAAAGCAATTGAAAGAAAA
ACAGGTGCGCGTGGTTTACGTTCAATCATAGAAGAATCGTTAATCGATATTATGTTTGAT
GTGCCTTCTAACGAAAATGTAACGAAGGTAGTTATTACAGCACAAACAATTAATGAAGAA
ACTGAACCAGAACTATACGACGCAGAAGGCAATTTAATTAATAATAGTAAAACATCAGCT
TAA60
120
180
240
300
360
420
480
540
600
660
720
780
840
900
960
1020
1080
1140
1200
1260
1263
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SAOUHSC_01778
- symbol: ClpX
- description: ATP-dependent protease ATP-binding subunit ClpX
- length: 420
- theoretical pI: 4.26448
- theoretical MW: 46297.2
- GRAVY: -0.265238
⊟Function[edit | edit source]
- TIGRFAM: Protein fate Degradation of proteins, peptides, and glycopeptides ATP-dependent Clp protease, ATP-binding subunit ClpX (TIGR00382; HMM-score: 660.9)Protein fate Protein folding and stabilization ATP-dependent Clp protease, ATP-binding subunit ClpX (TIGR00382; HMM-score: 660.9)and 25 moreProtein fate Protein folding and stabilization ATP-dependent protease HslVU, ATPase subunit (TIGR00390; HMM-score: 212.6)Protein fate Degradation of proteins, peptides, and glycopeptides ATP-dependent Clp protease ATP-binding subunit ClpA (TIGR02639; HMM-score: 46.1)Protein fate Protein folding and stabilization ATP-dependent chaperone protein ClpB (TIGR03346; HMM-score: 36.9)Cellular processes Pathogenesis type VI secretion ATPase, ClpV1 family (TIGR03345; HMM-score: 36.2)Protein fate Protein and peptide secretion and trafficking type VI secretion ATPase, ClpV1 family (TIGR03345; HMM-score: 36.2)AAA family ATPase, CDC48 subfamily (TIGR01243; HMM-score: 31.4)26S proteasome subunit P45 family (TIGR01242; HMM-score: 30)Cellular processes Cell division ATP-dependent metallopeptidase HflB (TIGR01241; EC 3.4.24.-; HMM-score: 27.4)Protein fate Degradation of proteins, peptides, and glycopeptides ATP-dependent metallopeptidase HflB (TIGR01241; EC 3.4.24.-; HMM-score: 27.4)DNA metabolism DNA replication, recombination, and repair Holliday junction DNA helicase RuvB (TIGR00635; EC 3.6.4.12; HMM-score: 25.4)Protein fate Degradation of proteins, peptides, and glycopeptides endopeptidase La (TIGR00763; EC 3.4.21.53; HMM-score: 23.8)Cellular processes Chemotaxis and motility flagellar biosynthesis protein FlhF (TIGR03499; HMM-score: 23.2)Unknown function General Mg chelatase-like protein (TIGR00368; HMM-score: 22.2)nicotinamide-nucleotide adenylyltransferase (TIGR01526; EC 2.7.7.1; HMM-score: 21.3)Cellular processes Other gas vesicle protein GvpN (TIGR02640; HMM-score: 20.1)Protein fate Degradation of proteins, peptides, and glycopeptides putative ATP-dependent protease (TIGR00764; HMM-score: 16.9)Hypothetical proteins Conserved TIGR00270 family protein (TIGR00270; HMM-score: 16.5)DNA metabolism DNA replication, recombination, and repair orc1/cdc6 family replication initiation protein (TIGR02928; HMM-score: 15.8)Purines, pyrimidines, nucleosides, and nucleotides Nucleotide and nucleoside interconversions cytidylate kinase (TIGR00017; EC 2.7.4.14; HMM-score: 14.5)Protein fate Protein and peptide secretion and trafficking type VII secretion AAA-ATPase EccA (TIGR03922; HMM-score: 13.1)Protein synthesis tRNA and rRNA base modification tRNA dimethylallyltransferase (TIGR00174; EC 2.5.1.75; HMM-score: 12.8)Transport and binding proteins Anions phosphate ABC transporter, ATP-binding protein (TIGR00972; EC 3.6.3.27; HMM-score: 12.8)thiol reductant ABC exporter, CydC subunit (TIGR02868; HMM-score: 12.8)Transport and binding proteins Anions sulfate ABC transporter, ATP-binding protein (TIGR00968; EC 3.6.3.25; HMM-score: 12.1)carbohydrate kinase, thermoresistant glucokinase family (TIGR01313; EC 2.7.1.-; HMM-score: 12.1)
- TheSEED :
- ATP-dependent Clp protease ATP-binding subunit ClpX
Protein Metabolism Protein degradation Proteasome bacterial ATP-dependent Clp protease ATP-binding subunit ClpXand 1 more - PFAM: P-loop_NTPase (CL0023) AAA_2; AAA domain (Cdc48 subfamily) (PF07724; HMM-score: 117.1)and 29 moreZn_Beta_Ribbon (CL0167) zf-C4_ClpX; ClpX C4-type zinc finger (PF06689; HMM-score: 67.7)P-loop_NTPase (CL0023) AAA; ATPase family associated with various cellular activities (AAA) (PF00004; HMM-score: 61.1)AAA_lid (CL0671) ClpB_D2-small; C-terminal, D2-small domain, of ClpB protein (PF10431; HMM-score: 47.6)P-loop_NTPase (CL0023) AAA_5; AAA domain (dynein-related subfamily) (PF07728; HMM-score: 28.2)AAA_22; AAA domain (PF13401; HMM-score: 27)RuvB_N; Holliday junction DNA helicase RuvB P-loop domain (PF05496; HMM-score: 26.2)Mg_chelatase; Magnesium chelatase, subunit ChlI (PF01078; HMM-score: 22.4)MCM; MCM P-loop domain (PF00493; HMM-score: 21.4)AAA_18; AAA domain (PF13238; HMM-score: 19.5)Sigma54_activ_2; Sigma-54 interaction domain (PF14532; HMM-score: 19.3)Sigma54_activat; Sigma-54 interaction domain (PF00158; HMM-score: 18.7)AAA_14; AAA domain (PF13173; HMM-score: 18)ABC_tran; ABC transporter (PF00005; HMM-score: 17.9)TsaE; Threonylcarbamoyl adenosine biosynthesis protein TsaE (PF02367; HMM-score: 17.9)IstB_IS21; IstB-like ATP binding protein (PF01695; HMM-score: 17.2)AAA_16; AAA ATPase domain (PF13191; HMM-score: 17)AAA_17; AAA domain (PF13207; HMM-score: 16.3)NACHT; NACHT domain (PF05729; HMM-score: 16)AAA_28; AAA domain (PF13521; HMM-score: 15.9)nSTAND3; Novel STAND NTPase 3 (PF20720; HMM-score: 15.5)ResIII; Type III restriction enzyme, res subunit (PF04851; HMM-score: 14.8)AAA_24; AAA domain (PF13479; HMM-score: 14.5)AAA_7; P-loop containing dynein motor region (PF12775; HMM-score: 14)Cytidylate_kin; Cytidylate kinase (PF02224; HMM-score: 13.8)DEAD; DEAD/DEAH box helicase (PF00270; HMM-score: 13.6)RNA_helicase; RNA helicase (PF00910; HMM-score: 13.5)AAA_33; AAA domain (PF13671; HMM-score: 13.1)PduV-EutP; Ethanolamine utilisation - propanediol utilisation (PF10662; HMM-score: 12.7)RING (CL0229) C1_2; C1 domain (PF03107; HMM-score: 11.3)
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors:
- effectors:
⊟Localization[edit | edit source]
- PSORTb: Cytoplasmic
- Cytoplasmic Score: 9.97
- Cytoplasmic Membrane Score: 0
- Cellwall Score: 0.01
- Extracellular Score: 0.02
- Internal Helices: 0
- DeepLocPro: Cytoplasmic
- Cytoplasmic Score: 0.9758
- Cytoplasmic Membrane Score: 0.0144
- Cell wall & surface Score: 0.0008
- Extracellular Score: 0.009
- LocateP: Intracellular
- Prediction by SwissProt Classification: Cytoplasmic
- Pathway Prediction: No pathway
- Intracellular possibility: 1
- Signal peptide possibility: -1
- N-terminally Anchored Score: 1
- Predicted Cleavage Site: No CleavageSite
- SignalP: no predicted signal peptide
- SP(Sec/SPI): 0.063176
- TAT(Tat/SPI): 0.001019
- LIPO(Sec/SPII): 0.00183
- predicted transmembrane helices (TMHMM): 0
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MFKFNEDEENLKCSFCGKDQDQVKKLVAGSGVYICNECIELCSEIVEEELAQNTSEAMTELPTPKEIMDHLNEYVIGQEKAKKSLAVAVYNHYKRIQQLGPKEDDVELQKSNIALIGPTGSGKTLLAQTLAKTLNVPFAIADATSLTEAGYVGDDVENILLRLIQAADFDIDKAEKGIIYVDEIDKIARKSENTSITRDVSGEGVQQALLKILEGTTASVPPQGGRKHPNQEMIQIDTTNILFILGGAFDGIEEVIKRRLGEKVIGFSSNEADKYDEQALLAQIRPEDLQAYGLIPEFIGRVPIVANLETLDVTALKNILTQPKNALVKQYTKMLELDDVDLEFTEEALSAISEKAIERKTGARGLRSIIEESLIDIMFDVPSNENVTKVVITAQTINEETEPELYDAEGNLINNSKTSA
⊟Experimental data[edit | edit source]
- experimentally validated: PeptideAtlas [1] [2]
- protein localization: data available for COL
- quantitative data / protein copy number per cell: data available for COL
- interaction partners:
SAOUHSC_02506 (rpsC) 30S ribosomal protein S3 [3] (data from MRSA252) SAOUHSC_02494 (rpsE) 30S ribosomal protein S5 [3] (data from MRSA252) SAOUHSC_00195 acetyl-CoA acetyltransferase [3] (data from MRSA252) SAOUHSC_00428 hypothetical protein [3] (data from MRSA252) SAOUHSC_02579 hypothetical protein [3] (data from MRSA252) SAOUHSC_02849 pyruvate oxidase [3] (data from MRSA252) SAOUHSC_02885 hypothetical protein [3] (data from MRSA252)
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
- MicrobesOnline: clpX < tigpredicted SigA promoter [4] : clpX < S703 < tig < SAOUHSC_01780 < SAOUHSC_01781 < SAOUHSC_01782 < rplT < rpmI < infC < S704 < S705
⊟Regulation[edit | edit source]
- regulator:
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: [4]
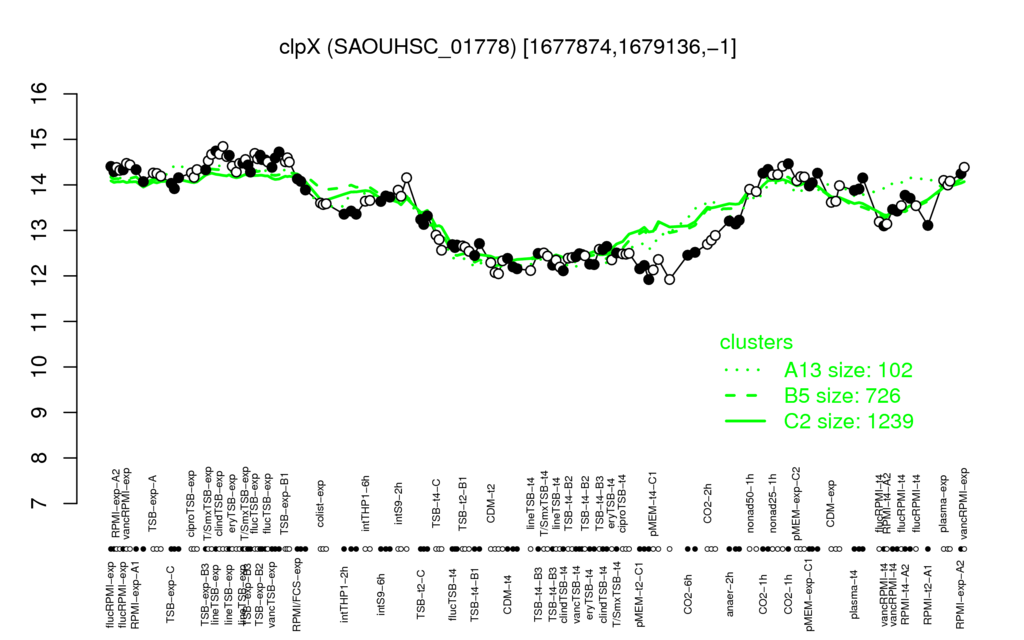 Multi-gene expression profiles
Multi-gene expression profiles
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
- Aureolib: no data available
⊟Protein stability[edit | edit source]
- half-life: no data available
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You can add further information about the gene and protein here. [edit]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ Maren Depke, Stephan Michalik, Alexander Rabe, Kristin Surmann, Lars Brinkmann, Nico Jehmlich, Jörg Bernhardt, Michael Hecker, Bernd Wollscheid, Zhi Sun, Robert L Moritz, Uwe Völker, Frank Schmidt
A peptide resource for the analysis of Staphylococcus aureus in host-pathogen interaction studies.
Proteomics: 2015, 15(21);3648-61
[PubMed:26224020] [WorldCat.org] [DOI] (I p) - ↑ Stephan Michalik, Maren Depke, Annette Murr, Manuela Gesell Salazar, Ulrike Kusebauch, Zhi Sun, Tanja C Meyer, Kristin Surmann, Henrike Pförtner, Petra Hildebrandt, Stefan Weiss, Laura Marcela Palma Medina, Melanie Gutjahr, Elke Hammer, Dörte Becher, Thomas Pribyl, Sven Hammerschmidt, Eric W Deutsch, Samuel L Bader, Michael Hecker, Robert L Moritz, Ulrike Mäder, Uwe Völker, Frank Schmidt
A global Staphylococcus aureus proteome resource applied to the in vivo characterization of host-pathogen interactions.
Sci Rep: 2017, 7(1);9718
[PubMed:28887440] [WorldCat.org] [DOI] (I e) - ↑ 3.0 3.1 3.2 3.3 3.4 3.5 3.6 Artem Cherkasov, Michael Hsing, Roya Zoraghi, Leonard J Foster, Raymond H See, Nikolay Stoynov, Jihong Jiang, Sukhbir Kaur, Tian Lian, Linda Jackson, Huansheng Gong, Rick Swayze, Emily Amandoron, Farhad Hormozdiari, Phuong Dao, Cenk Sahinalp, Osvaldo Santos-Filho, Peter Axerio-Cilies, Kendall Byler, William R McMaster, Robert C Brunham, B Brett Finlay, Neil E Reiner
Mapping the protein interaction network in methicillin-resistant Staphylococcus aureus.
J Proteome Res: 2011, 10(3);1139-50
[PubMed:21166474] [WorldCat.org] [DOI] (I p) - ↑ 4.0 4.1 4.2 Ulrike Mäder, Pierre Nicolas, Maren Depke, Jan Pané-Farré, Michel Debarbouille, Magdalena M van der Kooi-Pol, Cyprien Guérin, Sandra Dérozier, Aurelia Hiron, Hanne Jarmer, Aurélie Leduc, Stephan Michalik, Ewoud Reilman, Marc Schaffer, Frank Schmidt, Philippe Bessières, Philippe Noirot, Michael Hecker, Tarek Msadek, Uwe Völker, Jan Maarten van Dijl
Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions.
PLoS Genet: 2016, 12(4);e1005962
[PubMed:27035918] [WorldCat.org] [DOI] (I e)
