Jump to navigation
Jump to search
NCBI: 10-JUN-2013
⊟Summary[edit | edit source]
- organism: Staphylococcus aureus COL
- locus tag: SACOL0589 [new locus tag: SACOL_RS03055 ]
- pan locus tag?: SAUPAN002315000
- symbol: rpoC
- pan gene symbol?: rpoC
- synonym:
- product: DNA-directed RNA polymerase subunit beta'
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
- type: CDS
- locus tag: SACOL0589 [new locus tag: SACOL_RS03055 ]
- symbol: rpoC
- product: DNA-directed RNA polymerase subunit beta'
- replicon: chromosome
- strand: +
- coordinates: 610676..614299
- length: 3624
- essential: unknown other strains
⊟Accession numbers[edit | edit source]
- Gene ID: 3236179 NCBI
- RefSeq: YP_185475 NCBI
- BioCyc: see SACOL_RS03055
- MicrobesOnline: 912069 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421
481
541
601
661
721
781
841
901
961
1021
1081
1141
1201
1261
1321
1381
1441
1501
1561
1621
1681
1741
1801
1861
1921
1981
2041
2101
2161
2221
2281
2341
2401
2461
2521
2581
2641
2701
2761
2821
2881
2941
3001
3061
3121
3181
3241
3301
3361
3421
3481
3541
3601TTGATTGATGTAAATAATTTCCATTATATGAAAATAGGATTGGCTTCACCTGAAAAAATC
CGTTCTTGGTCTTTTGGTGAAGTTAAAAAACCTGAAACAATCAACTACCGTACATTAAAA
CCTGAAAAAGATGGTCTATTCTGTGAAAGAATTTTCGGACCTACAAAAGACTGGGAATGT
AGTTGTGGTAAATACAAACGTGTTCGCTACAAAGGCATGGTCTGTGACAGATGTGGAGTT
GAAGTAACTAAATCTAAAGTACGTCGTGAAAGAATGGGTCACATTGAACTTGCTGCTCCA
GTTTCTCACATTTGGTATTTCAAAGGTATACCAAGTCGTATGGGATTATTACTTGACATG
TCACCAAGAGCATTAGAAGAAGTTATTTACTTTGCTTCTTATGTTGTTGTAGATCCAGGT
CCAACTGGTTTAGAAAAGAAAACTTTATTATCTGAAGCTGAATTCAGAGATTATTATGAT
AAATACCCAGGTCAATTCGTTGCAAAAATGGGTGCAGAAGGTATTAAAGATTTACTTGAA
GAGATTGATCTTGACGAAGAACTTAAATTGTTACGCGATGAGTTGGAATCAGCTACTGGT
CAAAGACTTACTCGTGCAATTAAACGTTTAGAAGTTGTTGAATCATTCCGTAATTCAGGT
AACAAACCTTCATGGATGATTTTAGATGTACTTCCAATCATCCCACCAGAAATTCGTCCA
ATGGTTCAATTAGATGGTGGACGATTTGCAACAAGTGACTTAAACGACTTATACCGTCGT
GTAATTAATCGAAATAATCGTTTGAAACGTTTATTAGATTTAGGTGCACCTGGTATCATC
GTTCAAAACGAAAAACGTATGTTACAAGAAGCCGTTGACGCTTTAATTGATAATGGTCGT
CGTGGTCGTCCAGTTACTGGCCCAGGTAACCGTCCATTAAAATCTTTATCTCATATGTTA
AAAGGTAAACAAGGTCGTTTCCGTCAAAACTTACTTGGTAAACGTGTTGACTATTCAGGA
CGTTCAGTTATTGCAGTAGGTCCAAGCTTGAAAATGTACCAATGTGGTTTACCAAAAGAA
ATGGCACTTGAACTATTTAAACCATTCGTAATGAAAGAATTAGTTCAACGTGAAATTGCA
ACTAACATTAAAAATGCGAAGAGTAAAATCGAACGTATGGATGATGAAGTTTGGGACGTA
TTGGAAGAAGTAATTAGAGAACATCCTGTATTACTTAACCGTGCACCAACACTTCATAGA
CTTGGTATTCAAGCATTTGAACCAACTTTAGTTGAAGGTCGTGCGATTCGTCTACATCCA
CTTGTAACAACAGCTTATAACGCTGACTTTGACGGTGACCAAATGGCGGTTCACGTTCCT
TTATCAAAAGAGGCACAAGCTGAAGCAAGAATGTTGATGTTAGCAGCACAAAACATCTTG
AACCCTAAAGATGGTAAACCTGTAGTTACACCATCACAAGATATGGTACTTGGTAACTAT
TACCTTACTTTAGAAAGAAAAGATGCAGTAAATACAGGCGCAATCTTTAATAATACAAAT
GAAGTATTAAAAGCATATGCAAATGGCTTTGTACATTTACACACTAGAATTGGTGTACAT
GCAAGTTCGTTCAATAATCCAACATTTACTGAAGAACAAAACAAAAAGATTCTTGCTACG
TCAGTAGGTAAAATTATATTCAATGAAATCATTCCAGATTCATTTGCTTATATTAATGAA
CCTACGCAAGAAAACTTAGAAAGAAAGACACCAAACAGATATTTCATCGATCCTACAACT
TTAGGTGAAGGTGGATTAAAAGAATACTTTGAAAATGAAGAATTAATTGAACCTTTCAAC
AAAAAATTCTTAGGTAATATTATTGCAGAAGTATTCAACAGATTTAGCATCACTGATACA
TCAATGATGTTAGACCGTATGAAAGACTTAGGATTCAAATTCTCATCTAAAGCTGGTATT
ACAGTAGGTGTTGCTGATATCGTAGTATTACCTGATAAGCAACAAATACTTGATGAGCAT
GAAAAATTAGTCGACAGAATTACAAAACAATTCAACCGTGGTTTAATCACTGAAGAAGAA
AGATATAATGCAGTTGTTGAAATTTGGACAGATGCAAAAGATCAAATTCAAGGTGAATTG
ATGCAATCACTTGATAAAACTAACCCAATCTTCATGATGAGTGATTCAGGTGCCCGTGGT
AACGCATCTAACTTTACACAGTTAGCAGGTATGCGTGGATTGATGGCCGCACCATCTGGT
AAGATTATCGAATTACCAATCACATCTTCATTCCGTGAAGGTTTAACAGTACTTGAATAC
TTCATCTCAACTCACGGTGCACGTAAAGGTCTTGCCGATACAGCACTTAAAACAGCTGAC
TCAGGATATCTTACTCGTCGTCTTGTTGACGTGGCACAAGATGTTATTGTTCGTGAAGAA
GACTGTGGTACTGATAGAGGTTTATTAGTTTCTGATATTAAAGAAGGTACAGAAATGATT
GAACCATTTATCGAACGTATTGAAGGTCGTTATTCTAAAGAAACAATTCGTCATCCTGAA
ACTGATGAAATAATCATTCGTCCTGATGAATTAATTACACCTGAAATTGCTAAGAAAATT
ACAGATGCTGGTATTGAACAAATGTATATTCGCTCAGCATTTACTTGTAACGCACGACAT
GGTGTTTGTGAAAAATGTTACGGTAAAAACCTTGCTACTGGTGAAAAAGTTGAAGTTGGT
GAAGCAGTTGGTACAATTGCAGCCCAATCTATCGGTGAACCAGGTACACAGCTTACAATG
CGTACATTCCATACAGGTGGGGTAGCAGGTAGCGATATCACACAAGGTCTTCCTCGTATT
CAAGAGATTTTCGAAGCACGTAACCCTAAAGGTCAAGCGGTAATTACGGAAATCGAAGGT
GTCGTAGAAGATATTAAATTAGCAAAAGATAGACAACAAGAAATTGTTGTTAAAGGTGCT
AATGAAACAAGATCATACCTTGCTTCAGGTACTTCAAGAATTATTGTAGAAATCGGTCAA
CCAGTTCAACGTGGTGAAGTATTAACTGAAGGTTCTATTGAACCTAAGAATTACTTATCT
GTTGCTGGATTAAACGCGACTGAAAGCTACTTATTAAAAGAAGTACAAAAAGTTTACCGT
ATGCAAGGTGTAGAAATCGACGATAAACACGTTGAGGTTATGGTTCGACAAATGTTACGT
AAAGTTAGAATTATCGAAGCAGGTGATACGAAGTTATTACCAGGTTCATTAGTTGATATT
CATAACTTTACAGATGCAAATAGAGAAGCATTTAAACACCGTAAGCGTCCTGCAACAGCT
AAACCAGTATTACTTGGTATTACTAAAGCATCACTTGAAACAGAAAGTTTCTTATCTGCA
GCATCATTCCAAGAAACAACAAGAGTTCTTACAGATGCAGCAATTAAAGGTAAGCGTGAT
GACTTATTAGGTCTTAAAGAAAACGTAATTATTGGTAAGTTAATTCCAGCTGGTACTGGT
ATGAGACGTTATAGCGACGTAAAATACGAAAAAACAGCTAAACCAGTTGCAGAAGTTGAA
TCTCAAACTGAAGTAACGGAATAA60
120
180
240
300
360
420
480
540
600
660
720
780
840
900
960
1020
1080
1140
1200
1260
1320
1380
1440
1500
1560
1620
1680
1740
1800
1860
1920
1980
2040
2100
2160
2220
2280
2340
2400
2460
2520
2580
2640
2700
2760
2820
2880
2940
3000
3060
3120
3180
3240
3300
3360
3420
3480
3540
3600
3624
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SACOL0589 [new locus tag: SACOL_RS03055 ]
- symbol: RpoC
- description: DNA-directed RNA polymerase subunit beta'
- length: 1207
- theoretical pI: 6.93506
- theoretical MW: 135408
- GRAVY: -0.353024
⊟Function[edit | edit source]
- reaction: EC 2.7.7.6? ExPASyDNA-directed RNA polymerase Nucleoside triphosphate + RNA(n) = diphosphate + RNA(n+1)
- TIGRFAM: Transcription DNA-dependent RNA polymerase DNA-directed RNA polymerase, beta' subunit (TIGR02386; EC 2.7.7.6; HMM-score: 1884.2)and 4 moreDNA-directed RNA polymerase, gamma subunit (TIGR02387; EC 2.7.7.6; HMM-score: 850.1)Transcription DNA-dependent RNA polymerase DNA-directed RNA polymerase, beta subunit (TIGR02388; EC 2.7.7.6; HMM-score: 645.3)DNA-directed RNA polymerase subunit A' (TIGR02390; EC 2.7.7.6; HMM-score: 322.2)Transcription DNA-dependent RNA polymerase DNA-directed RNA polymerase, subunit A (TIGR02389; EC 2.7.7.6; HMM-score: 115)
- TheSEED :
- DNA-directed RNA polymerase beta' subunit (EC 2.7.7.6)
RNA Metabolism Transcription RNA polymerase bacterial DNA-directed RNA polymerase beta' subunit (EC 2.7.7.6)and 1 more - PFAM: no clan defined RNA_pol_Rpb1_1; RNA polymerase Rpb1, domain 1 (PF04997; HMM-score: 286.9)RNA_pol_Rpb1_5; RNA polymerase Rpb1, domain 5 (PF04998; HMM-score: 233.6)and 4 moreRNA_pol_Rpb1_2; RNA polymerase Rpb1, domain 2 (PF00623; HMM-score: 142.8)RNA_pol_Rpb1_3; RNA polymerase Rpb1, domain 3 (PF04983; HMM-score: 137.6)RNA_pol_Rpb1_4; RNA polymerase Rpb1, domain 4 (PF05000; HMM-score: 63.7)Hybrid (CL0105) Peptidase_M23; Peptidase family M23 (PF01551; HMM-score: 17.5)
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors:
- effectors:
⊟Localization[edit | edit source]
- PSORTb: Cytoplasmic
- Cytoplasmic Score: 9.97
- Cytoplasmic Membrane Score: 0
- Cellwall Score: 0.01
- Extracellular Score: 0.02
- Internal Helices: 0
- DeepLocPro: Cytoplasmic
- Cytoplasmic Score: 0.8613
- Cytoplasmic Membrane Score: 0.0431
- Cell wall & surface Score: 0.0032
- Extracellular Score: 0.0924
- LocateP: Intracellular
- Prediction by SwissProt Classification: Cytoplasmic
- Pathway Prediction: No pathway
- Intracellular possibility: 1
- Signal peptide possibility: -1
- N-terminally Anchored Score: 1
- Predicted Cleavage Site: No CleavageSite
- SignalP: no predicted signal peptide
- SP(Sec/SPI): 0.052911
- TAT(Tat/SPI): 0.002526
- LIPO(Sec/SPII): 0.005436
- predicted transmembrane helices (TMHMM): 0
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MIDVNNFHYMKIGLASPEKIRSWSFGEVKKPETINYRTLKPEKDGLFCERIFGPTKDWECSCGKYKRVRYKGMVCDRCGVEVTKSKVRRERMGHIELAAPVSHIWYFKGIPSRMGLLLDMSPRALEEVIYFASYVVVDPGPTGLEKKTLLSEAEFRDYYDKYPGQFVAKMGAEGIKDLLEEIDLDEELKLLRDELESATGQRLTRAIKRLEVVESFRNSGNKPSWMILDVLPIIPPEIRPMVQLDGGRFATSDLNDLYRRVINRNNRLKRLLDLGAPGIIVQNEKRMLQEAVDALIDNGRRGRPVTGPGNRPLKSLSHMLKGKQGRFRQNLLGKRVDYSGRSVIAVGPSLKMYQCGLPKEMALELFKPFVMKELVQREIATNIKNAKSKIERMDDEVWDVLEEVIREHPVLLNRAPTLHRLGIQAFEPTLVEGRAIRLHPLVTTAYNADFDGDQMAVHVPLSKEAQAEARMLMLAAQNILNPKDGKPVVTPSQDMVLGNYYLTLERKDAVNTGAIFNNTNEVLKAYANGFVHLHTRIGVHASSFNNPTFTEEQNKKILATSVGKIIFNEIIPDSFAYINEPTQENLERKTPNRYFIDPTTLGEGGLKEYFENEELIEPFNKKFLGNIIAEVFNRFSITDTSMMLDRMKDLGFKFSSKAGITVGVADIVVLPDKQQILDEHEKLVDRITKQFNRGLITEEERYNAVVEIWTDAKDQIQGELMQSLDKTNPIFMMSDSGARGNASNFTQLAGMRGLMAAPSGKIIELPITSSFREGLTVLEYFISTHGARKGLADTALKTADSGYLTRRLVDVAQDVIVREEDCGTDRGLLVSDIKEGTEMIEPFIERIEGRYSKETIRHPETDEIIIRPDELITPEIAKKITDAGIEQMYIRSAFTCNARHGVCEKCYGKNLATGEKVEVGEAVGTIAAQSIGEPGTQLTMRTFHTGGVAGSDITQGLPRIQEIFEARNPKGQAVITEIEGVVEDIKLAKDRQQEIVVKGANETRSYLASGTSRIIVEIGQPVQRGEVLTEGSIEPKNYLSVAGLNATESYLLKEVQKVYRMQGVEIDDKHVEVMVRQMLRKVRIIEAGDTKLLPGSLVDIHNFTDANREAFKHRKRPATAKPVLLGITKASLETESFLSAASFQETTRVLTDAAIKGKRDDLLGLKENVIIGKLIPAGTGMRRYSDVKYEKTAKPVAEVESQTEVTE
⊟Experimental data[edit | edit source]
- experimentally validated: PeptideAtlas
- protein localization: Cytoplasmic [1] [2] [3] [4]
- quantitative data / protein copy number per cell: 3043 [5]
- interaction partners:
SACOL2657 (arcA) arginine deiminase [6] (data from MRSA252) SACOL2130 (deoD) purine nucleoside phosphorylase [6] (data from MRSA252) SACOL1587 (efp) elongation factor P [6] (data from MRSA252) SACOL0593 (fusA) elongation factor G [6] (data from MRSA252) SACOL1734 (gapA2) glyceraldehyde 3-phosphate dehydrogenase 2 [6] (data from MRSA252) SACOL2016 (groEL) chaperonin GroEL [6] (data from MRSA252) SACOL0006 (gyrA) DNA gyrase, A subunit [6] (data from MRSA252) SACOL1206 (ileS) isoleucyl-tRNA synthetase [6] (data from MRSA252) SACOL0240 (ispD) 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase [6] (data from MRSA252) SACOL2224 (rplF) 50S ribosomal protein L6 [6] (data from MRSA252) SACOL1257 (rplS) 50S ribosomal protein L19 [6] (data from MRSA252) SACOL2234 (rplV) 50S ribosomal protein L22 [6] (data from MRSA252) SACOL0592 (rpsG) 30S ribosomal protein S7 [6] (data from MRSA252) SACOL2230 (rpsQ) 30S ribosomal protein S17 [6] (data from MRSA252) SACOL0594 (tuf) elongation factor Tu [6] (data from MRSA252) SACOL0731 LysR family transcriptional regulator [6] (data from MRSA252) SACOL1503 hypothetical protein [6] (data from MRSA252) SACOL1759 universal stress protein [6] (data from MRSA252) SACOL2293 NAD/NADP octopine/nopaline dehydrogenase [6] (data from MRSA252)
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
⊟Regulation[edit | edit source]
- regulator:
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: data available for NCTC8325
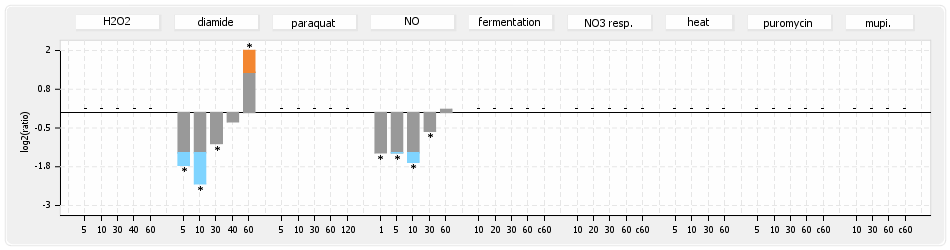
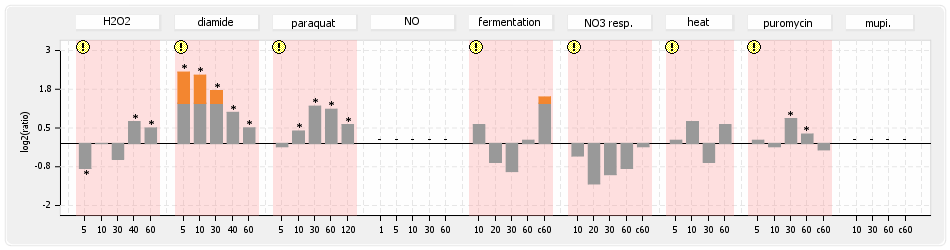
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
⊟Protein stability[edit | edit source]
- half-life: 110.53 h [7]
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You can add further information about the gene and protein here. [edit]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ Dörte Becher, Kristina Hempel, Susanne Sievers, Daniela Zühlke, Jan Pané-Farré, Andreas Otto, Stephan Fuchs, Dirk Albrecht, Jörg Bernhardt, Susanne Engelmann, Uwe Völker, Jan Maarten van Dijl, Michael Hecker
A proteomic view of an important human pathogen--towards the quantification of the entire Staphylococcus aureus proteome.
PLoS One: 2009, 4(12);e8176
[PubMed:19997597] [WorldCat.org] [DOI] (I e) - ↑ Kristina Hempel, Jan Pané-Farré, Andreas Otto, Susanne Sievers, Michael Hecker, Dörte Becher
Quantitative cell surface proteome profiling for SigB-dependent protein expression in the human pathogen Staphylococcus aureus via biotinylation approach.
J Proteome Res: 2010, 9(3);1579-90
[PubMed:20108986] [WorldCat.org] [DOI] (I p) - ↑ Kristina Hempel, Florian-Alexander Herbst, Martin Moche, Michael Hecker, Dörte Becher
Quantitative proteomic view on secreted, cell surface-associated, and cytoplasmic proteins of the methicillin-resistant human pathogen Staphylococcus aureus under iron-limited conditions.
J Proteome Res: 2011, 10(4);1657-66
[PubMed:21323324] [WorldCat.org] [DOI] (I p) - ↑ Andreas Otto, Jan Maarten van Dijl, Michael Hecker, Dörte Becher
The Staphylococcus aureus proteome.
Int J Med Microbiol: 2014, 304(2);110-20
[PubMed:24439828] [WorldCat.org] [DOI] (I p) - ↑ Daniela Zühlke, Kirsten Dörries, Jörg Bernhardt, Sandra Maaß, Jan Muntel, Volkmar Liebscher, Jan Pané-Farré, Katharina Riedel, Michael Lalk, Uwe Völker, Susanne Engelmann, Dörte Becher, Stephan Fuchs, Michael Hecker
Costs of life - Dynamics of the protein inventory of Staphylococcus aureus during anaerobiosis.
Sci Rep: 2016, 6;28172
[PubMed:27344979] [WorldCat.org] [DOI] (I e) - ↑ 6.00 6.01 6.02 6.03 6.04 6.05 6.06 6.07 6.08 6.09 6.10 6.11 6.12 6.13 6.14 6.15 6.16 6.17 6.18 Artem Cherkasov, Michael Hsing, Roya Zoraghi, Leonard J Foster, Raymond H See, Nikolay Stoynov, Jihong Jiang, Sukhbir Kaur, Tian Lian, Linda Jackson, Huansheng Gong, Rick Swayze, Emily Amandoron, Farhad Hormozdiari, Phuong Dao, Cenk Sahinalp, Osvaldo Santos-Filho, Peter Axerio-Cilies, Kendall Byler, William R McMaster, Robert C Brunham, B Brett Finlay, Neil E Reiner
Mapping the protein interaction network in methicillin-resistant Staphylococcus aureus.
J Proteome Res: 2011, 10(3);1139-50
[PubMed:21166474] [WorldCat.org] [DOI] (I p) - ↑ Stephan Michalik, Jörg Bernhardt, Andreas Otto, Martin Moche, Dörte Becher, Hanna Meyer, Michael Lalk, Claudia Schurmann, Rabea Schlüter, Holger Kock, Ulf Gerth, Michael Hecker
Life and death of proteins: a case study of glucose-starved Staphylococcus aureus.
Mol Cell Proteomics: 2012, 11(9);558-70
[PubMed:22556279] [WorldCat.org] [DOI] (I p)