Jump to navigation
Jump to search
NCBI: 03-AUG-2016
⊟Summary[edit | edit source]
- organism: Staphylococcus aureus NCTC8325
- locus tag: SAOUHSC_00258
- pan locus tag?: SAUPAN001176000
- symbol: SAOUHSC_00258
- pan gene symbol?: esaA
- synonym:
- product: hypothetical protein
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
- type: CDS
- locus tag: SAOUHSC_00258
- symbol: SAOUHSC_00258
- product: hypothetical protein
- replicon: chromosome
- strand: +
- coordinates: 276308..279337
- length: 3030
- essential: no DEG other strains
⊟Accession numbers[edit | edit source]
- Gene ID: 3919200 NCBI
- RefSeq: YP_498852 NCBI
- BioCyc: G1I0R-240 BioCyc
- MicrobesOnline: 1288746 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421
481
541
601
661
721
781
841
901
961
1021
1081
1141
1201
1261
1321
1381
1441
1501
1561
1621
1681
1741
1801
1861
1921
1981
2041
2101
2161
2221
2281
2341
2401
2461
2521
2581
2641
2701
2761
2821
2881
2941
3001ATGAAAAAGAAAAATTGGATTTATGCATTAATTGTCACTTTAATTATTATAATTGCCATA
GTTAGTATGATATTTTTTGTTCAAACAAAATATGGAGATCAATCAGAAAAAGGATCCCAA
AGTGTAAGTAATAAAAATAATAAAATACATATCGCAATTGTTAACGAGGATCAACCAACG
ACATATAACGGTAAAAAAGTTGAGCTGGGTCAAGCATTTATTAAAAGGTTAGCAAATGAG
AAAAACTATAAATTTGAAACAGTAACAAGAAACGTTGCTGAGTCTGGTTTGAAAAATGGT
GGATACCAAGTCATGATTGTTATCCCAGAAAACTTTTCAAAATTGGCAATGCAATTAGAC
GCTAAAACACCATCGAAAATATCGCTACAGTATAAAACAGCTGTAGGACAAAAAGAAGAA
GTAGCTAAAAACACAGAAAAAGTTGTAAGTAATGTACTTAACGACTTTAACAAAAACTTA
GTCGAAATTTATTTAACAAGCATCATTGATAATTTACATAATGCACAAAAAAATGTTGGC
GCTATTATGACGCGTGAACATGGTGTGAATAGTAAATTCTCGAATTACTTATTAAATCCA
ATTAACGACTTCCCGGAATTATTTACAGATACGCTTGTAAATTCAATTTCTGCAAACAAA
GACATTACAAAATGGTTCCAAACATACAATAAATCATTATTGAGTGCGAATTCAGATACG
TTCAGAGTGAACACAGATTATAATGTTTCGACTTTAATTGAAAAACAAAATTCATTATTT
GACGAGCACAATACAGCGATGGATAAAATGTTACAAGATTATAAATCGCAAAAAGATAGC
GTGGAACTTGATAACTATATCAATGCATTAAAACAGATGGACAGCCAAATTGATCAACAA
TCAAGTATGCAAGATACAGGTAAAGAAGAATATAAACAAACTGTTAAAGAAAACTTAGAT
AAATTAAGAGAAATCATTCAATCACAAGAGTCACCATTTTCAAAAGGTATGATTGAAGAC
TATCGTAAGCAATTAACAGAATCACTGCAAGATGAGCTTGCAAATAACAAAGACTTACAA
GATGCGCTAAATAGCATTAAAATGAACAATGCTCAATTCGCTGAAAACTTAGAGAAACAA
CTTCATGATGATATTGTCAAAGAACCTGATACAGATACAACATTTATCTATAACATGTCT
AAACAAGACTTTATAGCTGCAGGTTTAAATGAGGATGAAGCTAATAAATACGAAGCAATT
GTCAAAGAAGCAAAACGTTATAAAAATGAATATAATTTGAAAAAACCGTTAGCAGAACAC
ATTAATTTAACAGATTACGATAACCAAGTTGCGCAAGACACAAGTAGTTTGATTAATGAT
GGTGTCAAAGTGCAACGTACTGAAACGATTAAAAGTAATGATATTAATCAATTAACTGTT
GCAACAGATCCTCATTTTAATTTTGAAGGCGACATTAAAATTAATGGTAAAAAATATGAC
ATTAAGGATCAAAGTGTTCAACTCGATACATCTAACAAGGAATATAAAGTTGAAGTCAAT
GGCGTTGCTAAATTGAAAAAGGATGCTGAGAAAGATTTCTTAAAAGATAAAACAATGCAT
TTACAATTGTTATTTGGACAAGCAAATCGTCAAGATGAACCAAATGATAAGAAAGCAACG
AGTGTTGTGGATGTAACATTGAATCATAACCTTGATGGTCGCTTATCGAAAGATGCATTA
AGCCAGCAATTGAGTGCATTATCTAGGTTTGATGCGCATTATAAAATGTACACAGATACA
AAAGGCAGAGAAGATAAACCATTCGACAACAAACGTTTAATTGATATGATGGTTGACCAA
GTTATCAATGACATGGAAAGTTTCAAAGACGATAAAGTAGCTGTGTTACATCAAATTGAT
TCAATGGAAGAAAACTCAGACAAACTGATTGATGACATTTTAAATAACAAAAAGAATACA
ACAAAAAATAAAGAAGATATTTCCAAGCTGATTGATCAGTTAGAAAACGTTAAAAAGACT
TTTGCTGAAGAGCCACAAGAACCAAAAATTGATAAAGGCAAAAATGATGAATTTAATACG
ATGTCTTCAAATTTAGATAAAGAAATTAGTAGAATTTCTGAAAAGAGTACGCAATTGCTA
TCAGATACACAAGAATCAAAATCAATTGCAGATTCTGTTAGTGGCCAATTAAATCAAGTC
GACAATAATGTGAATAAGCTACATGCGACAGGTCGAGCATTAGGCGTAAGAGCTAACGAT
TTGAATCGTCAAATGGCTAAAAACGATAAAGATAATGAGTTGTTCGCTAAAGAATTTAAA
AAAGTATTACAAAATTCTAAAGATGGCGACAGACAAAACCAAGCATTAAAAGCATTTATG
AGTAATCCGGTTCAAAAGAAAAACTTAGAAAATGTTTTAGCTAATAATGGTAATACAGAC
GTGATTTCACCGACATTATTCGTATTATTGATGTATTTACTATCAATGATTACAGCATAT
ATTTTCTATAGTTATGAACGTGCCAAAGGACAAATGAATTTCATTAAAGATGATTATAGT
AGTAAAAACCATCTTTGGAATAATGTCATTACGTCAGGTGTTATTGGTACAACTGGTTTG
GTAGAAGGGTTAATTGTCGGTTTAATTGCAATGAATAAGTTCCATGTATTAGCTGGCTAT
AGAGCGAAATTCATCTTAATGGTGATTTTAACTATGATGGTCTTCGTACTTATTAATACG
TATTTACTAAGACAGGTAAAATCTATCGGTATGTTCTTAATGATTGCTGCATTGGGTCTA
TACTTTGTAGCTATGAATAATTTGAAAGCAGCTGGACAAGGTGTGACTAATAAAATTTCA
CCATTGTCTTATATCGATAACATGTTCTTCAATTATTTAAATGCAGAGCATCCTATAGGC
TTGGTGCTAGTAATATTAACAGTACTTGTGATTATTGGCTTTGTACTGAACATGTTTATA
AAACACTTTAAGAAAGAGAGATTAATCTAA60
120
180
240
300
360
420
480
540
600
660
720
780
840
900
960
1020
1080
1140
1200
1260
1320
1380
1440
1500
1560
1620
1680
1740
1800
1860
1920
1980
2040
2100
2160
2220
2280
2340
2400
2460
2520
2580
2640
2700
2760
2820
2880
2940
3000
3030
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SAOUHSC_00258
- symbol: SAOUHSC_00258
- description: hypothetical protein
- length: 1009
- theoretical pI: 6.53987
- theoretical MW: 114825
- GRAVY: -0.507235
⊟Function[edit | edit source]
- TIGRFAM: type VII secretion protein EsaA (TIGR03929; HMM-score: 239.7)and 2 moreYhgE/Pip N-terminal domain (TIGR03061; HMM-score: 88.6)KxYKxGKxW signal peptide (TIGR03715; HMM-score: 9.1)
- TheSEED :
- Putative secretion accessory protein EsaA/YueB
- PFAM: MFS (CL0015) OATP; Organic Anion Transporter Polypeptide (OATP) family (PF03137; HMM-score: 15.1)no clan defined DUF3149; Protein of unknown function (DUF3149) (PF11346; HMM-score: 14.7)NDUF_B6; NADH:ubiquinone oxidoreductase, NDUFB6/B17 subunit (PF09782; HMM-score: 13.2)and 6 moreDUF1206; Domain of Unknown Function (DUF1206) (PF06724; HMM-score: 10.8)PTPA; Phosphotyrosyl phosphate activator (PTPA) protein (PF03095; HMM-score: 9.7)DUF4093; Domain of unknown function (DUF4093) (PF13331; HMM-score: 8.4)TPR (CL0020) COG4_N; Conserved oligomeric Golgi complex subunit 4, N-terminal (PF20663; HMM-score: 8)no clan defined KxYKxGKxW_sig; KxYKxGKxW signal peptide (PF19258; HMM-score: 7.7)TPR (CL0020) HEAT_EF3_N; EF3 N-terminal HEAT repeat (PF24987; HMM-score: 6.5)
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors:
- effectors:
⊟Localization[edit | edit source]
- PSORTb: Cytoplasmic Membrane
- Cytoplasmic Score: 0
- Cytoplasmic Membrane Score: 10
- Cellwall Score: 0
- Extracellular Score: 0
- Internal Helices: 6
- DeepLocPro: Cytoplasmic Membrane
- Cytoplasmic Score: 0.0033
- Cytoplasmic Membrane Score: 0.9068
- Cell wall & surface Score: 0.0181
- Extracellular Score: 0.0719
- LocateP: Multi-transmembrane
- Prediction by SwissProt Classification: Membrane
- Pathway Prediction: Sec-(SPI)
- Intracellular possibility: 0.17
- Signal peptide possibility: 0
- N-terminally Anchored Score: -1
- Predicted Cleavage Site: No CleavageSite
- SignalP: Signal peptide SP(Sec/SPI) length 32 aa
- SP(Sec/SPI): 0.475242
- TAT(Tat/SPI): 0.000917
- LIPO(Sec/SPII): 0.051651
- Cleavage Site: CS pos: 32-33. KYG-DQ. Pr: 0.0925
- predicted transmembrane helices (TMHMM): 6
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MKKKNWIYALIVTLIIIIAIVSMIFFVQTKYGDQSEKGSQSVSNKNNKIHIAIVNEDQPTTYNGKKVELGQAFIKRLANEKNYKFETVTRNVAESGLKNGGYQVMIVIPENFSKLAMQLDAKTPSKISLQYKTAVGQKEEVAKNTEKVVSNVLNDFNKNLVEIYLTSIIDNLHNAQKNVGAIMTREHGVNSKFSNYLLNPINDFPELFTDTLVNSISANKDITKWFQTYNKSLLSANSDTFRVNTDYNVSTLIEKQNSLFDEHNTAMDKMLQDYKSQKDSVELDNYINALKQMDSQIDQQSSMQDTGKEEYKQTVKENLDKLREIIQSQESPFSKGMIEDYRKQLTESLQDELANNKDLQDALNSIKMNNAQFAENLEKQLHDDIVKEPDTDTTFIYNMSKQDFIAAGLNEDEANKYEAIVKEAKRYKNEYNLKKPLAEHINLTDYDNQVAQDTSSLINDGVKVQRTETIKSNDINQLTVATDPHFNFEGDIKINGKKYDIKDQSVQLDTSNKEYKVEVNGVAKLKKDAEKDFLKDKTMHLQLLFGQANRQDEPNDKKATSVVDVTLNHNLDGRLSKDALSQQLSALSRFDAHYKMYTDTKGREDKPFDNKRLIDMMVDQVINDMESFKDDKVAVLHQIDSMEENSDKLIDDILNNKKNTTKNKEDISKLIDQLENVKKTFAEEPQEPKIDKGKNDEFNTMSSNLDKEISRISEKSTQLLSDTQESKSIADSVSGQLNQVDNNVNKLHATGRALGVRANDLNRQMAKNDKDNELFAKEFKKVLQNSKDGDRQNQALKAFMSNPVQKKNLENVLANNGNTDVISPTLFVLLMYLLSMITAYIFYSYERAKGQMNFIKDDYSSKNHLWNNVITSGVIGTTGLVEGLIVGLIAMNKFHVLAGYRAKFILMVILTMMVFVLINTYLLRQVKSIGMFLMIAALGLYFVAMNNLKAAGQGVTNKISPLSYIDNMFFNYLNAEHPIGLVLVILTVLVIIGFVLNMFIKHFKKERLI
⊟Experimental data[edit | edit source]
- experimentally validated: PeptideAtlas [1] [2]
- protein localization: data available for COL
- quantitative data / protein copy number per cell:
- interaction partners:
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
⊟Regulation[edit | edit source]
- regulator:
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: [3]
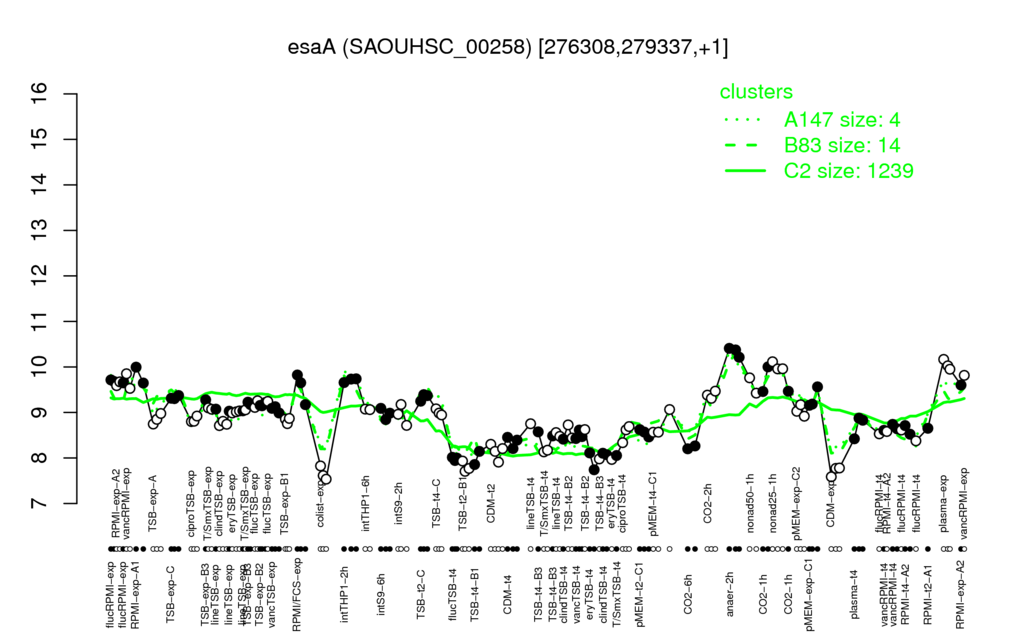 Multi-gene expression profiles
Multi-gene expression profiles
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
- Aureolib: no data available
⊟Protein stability[edit | edit source]
- half-life: no data available
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You can add further information about the gene and protein here. [edit]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ Maren Depke, Stephan Michalik, Alexander Rabe, Kristin Surmann, Lars Brinkmann, Nico Jehmlich, Jörg Bernhardt, Michael Hecker, Bernd Wollscheid, Zhi Sun, Robert L Moritz, Uwe Völker, Frank Schmidt
A peptide resource for the analysis of Staphylococcus aureus in host-pathogen interaction studies.
Proteomics: 2015, 15(21);3648-61
[PubMed:26224020] [WorldCat.org] [DOI] (I p) - ↑ Stephan Michalik, Maren Depke, Annette Murr, Manuela Gesell Salazar, Ulrike Kusebauch, Zhi Sun, Tanja C Meyer, Kristin Surmann, Henrike Pförtner, Petra Hildebrandt, Stefan Weiss, Laura Marcela Palma Medina, Melanie Gutjahr, Elke Hammer, Dörte Becher, Thomas Pribyl, Sven Hammerschmidt, Eric W Deutsch, Samuel L Bader, Michael Hecker, Robert L Moritz, Ulrike Mäder, Uwe Völker, Frank Schmidt
A global Staphylococcus aureus proteome resource applied to the in vivo characterization of host-pathogen interactions.
Sci Rep: 2017, 7(1);9718
[PubMed:28887440] [WorldCat.org] [DOI] (I e) - ↑ Ulrike Mäder, Pierre Nicolas, Maren Depke, Jan Pané-Farré, Michel Debarbouille, Magdalena M van der Kooi-Pol, Cyprien Guérin, Sandra Dérozier, Aurelia Hiron, Hanne Jarmer, Aurélie Leduc, Stephan Michalik, Ewoud Reilman, Marc Schaffer, Frank Schmidt, Philippe Bessières, Philippe Noirot, Michael Hecker, Tarek Msadek, Uwe Völker, Jan Maarten van Dijl
Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions.
PLoS Genet: 2016, 12(4);e1005962
[PubMed:27035918] [WorldCat.org] [DOI] (I e)
