Jump to navigation
Jump to search
NCBI: 03-AUG-2016
⊟Summary[edit | edit source]
- organism: Staphylococcus aureus NCTC8325
- locus tag: SAOUHSC_00970
- pan locus tag?: SAUPAN003225000
- symbol: SAOUHSC_00970
- pan gene symbol?: —
- synonym:
- product: ABC transporter ATP-binding protein
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
- type: CDS
- locus tag: SAOUHSC_00970
- symbol: SAOUHSC_00970
- product: ABC transporter ATP-binding protein
- replicon: chromosome
- strand: +
- coordinates: 945614..946255
- length: 642
- essential: no DEG other strains
⊟Accession numbers[edit | edit source]
- Gene ID: 3920115 NCBI
- RefSeq: YP_499523 NCBI
- BioCyc: G1I0R-914 BioCyc
- MicrobesOnline: 1289436 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421
481
541
601ATGATAGAATTAAAAGATTTAACCATACAAAAAGGTAATACACATATTTTGAAGAAACTT
AATTTGAAATTTCAATGTGGAAAATCATATGCACTTATTGGTAAAAGTGGATGCGGTAAA
TCTACATTATTAAATACTATTGCTGGACTTGAAAAAACAGGAAAACAATATGTCTATTTT
AATGGTCAATTAGAACAATTTAAATCTAATTTTTACAGAGATAAATTAGGATATTTATTT
CAAAATTATGGATTAATCGATAATTTGACAGTAAATGAAAATTTAGATATTGGATTAGCA
TATAAAAAAATAAGTAAGAAAGAAAAAGAACAAATAAAGATACGTTATATAGAACAGTTT
GGTCTGTCAAACAGTTTAAAAAGAAAAGTTCATACGCTAAGTGGAGGTGAACAACAACGT
GTCGCTTTAATTAGAATGATGTTAAAAGATCCGATTGTTATGTTAGCTGATGAACCAACG
GGTGCGTTAGATCCTAAAACAGGACAGATGATTATTCAATCATTATTTGGTTTGGTCGAT
GAAAATAAAGTGCTGATTTTAGCAACACATGATATGGCTATTGCAAATCAATGTGACGAA
ATAATAGATTTAGAACAGTATAGAAAAGTAGCATCTATGTGA60
120
180
240
300
360
420
480
540
600
642
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SAOUHSC_00970
- symbol: SAOUHSC_00970
- description: ABC transporter ATP-binding protein
- length: 213
- theoretical pI: 9.562
- theoretical MW: 24113
- GRAVY: -0.26338
⊟Function[edit | edit source]
- TIGRFAM: Cellular processes Toxin production and resistance putative bacteriocin export ABC transporter, lactococcin 972 group (TIGR03608; HMM-score: 267.3)Transport and binding proteins Unknown substrate putative bacteriocin export ABC transporter, lactococcin 972 group (TIGR03608; HMM-score: 267.3)and 78 moreProtein fate Protein and peptide secretion and trafficking lipoprotein releasing system, ATP-binding protein (TIGR02211; EC 3.6.3.-; HMM-score: 142.9)Cellular processes Cell division cell division ATP-binding protein FtsE (TIGR02673; HMM-score: 140.1)thiol reductant ABC exporter, CydD subunit (TIGR02857; HMM-score: 134.7)Transport and binding proteins Anions phosphonate ABC transporter, ATP-binding protein (TIGR02315; EC 3.6.3.28; HMM-score: 133.4)Transport and binding proteins Amino acids, peptides and amines putative 2-aminoethylphosphonate ABC transporter, ATP-binding protein (TIGR03265; HMM-score: 129.1)ABC exporter ATP-binding subunit, DevA family (TIGR02982; HMM-score: 128.4)Transport and binding proteins Anions sulfate ABC transporter, ATP-binding protein (TIGR00968; EC 3.6.3.25; HMM-score: 127.4)Transport and binding proteins Other thiamine ABC transporter, ATP-binding protein (TIGR01277; EC 3.6.3.-; HMM-score: 115.8)Transport and binding proteins Anions nitrate ABC transporter, ATP-binding proteins C and D (TIGR01184; HMM-score: 112.7)Transport and binding proteins Other nitrate ABC transporter, ATP-binding proteins C and D (TIGR01184; HMM-score: 112.7)Transport and binding proteins Anions phosphate ABC transporter, ATP-binding protein (TIGR00972; EC 3.6.3.27; HMM-score: 111.7)Transport and binding proteins Anions molybdate ABC transporter, ATP-binding protein (TIGR02142; EC 3.6.3.29; HMM-score: 111.4)Transport and binding proteins Carbohydrates, organic alcohols, and acids ABC transporter, ATP-binding subunit, PQQ-dependent alcohol dehydrogenase system (TIGR03864; HMM-score: 111.1)Protein fate Protein and peptide secretion and trafficking heme ABC exporter, ATP-binding protein CcmA (TIGR01189; EC 3.6.3.41; HMM-score: 111)Transport and binding proteins Other heme ABC exporter, ATP-binding protein CcmA (TIGR01189; EC 3.6.3.41; HMM-score: 111)Transport and binding proteins Unknown substrate energy-coupling factor transporter ATPase (TIGR04520; EC 3.6.3.-; HMM-score: 110)2-aminoethylphosphonate ABC transport system, ATP-binding component PhnT (TIGR03258; HMM-score: 109.8)Transport and binding proteins Unknown substrate energy-coupling factor transporter ATPase (TIGR04521; EC 3.6.3.-; HMM-score: 107.3)Transport and binding proteins Amino acids, peptides and amines glycine betaine/L-proline transport ATP binding subunit (TIGR01186; HMM-score: 105.3)Transport and binding proteins Amino acids, peptides and amines urea ABC transporter, ATP-binding protein UrtE (TIGR03410; HMM-score: 105.2)Cellular processes Pathogenesis type I secretion system ATPase (TIGR03375; HMM-score: 104.7)Protein fate Protein and peptide secretion and trafficking type I secretion system ATPase (TIGR03375; HMM-score: 104.7)thiol reductant ABC exporter, CydC subunit (TIGR02868; HMM-score: 104.2)Transport and binding proteins Amino acids, peptides and amines polyamine ABC transporter, ATP-binding protein (TIGR01187; EC 3.6.3.31; HMM-score: 104.1)Transport and binding proteins Other daunorubicin resistance ABC transporter, ATP-binding protein (TIGR01188; HMM-score: 102.4)Transport and binding proteins Other pigment precursor permease (TIGR00955; HMM-score: 102.3)Transport and binding proteins Cations and iron carrying compounds cobalt ABC transporter, ATP-binding protein (TIGR01166; HMM-score: 96.9)D-methionine ABC transporter, ATP-binding protein (TIGR02314; EC 3.6.3.-; HMM-score: 95.6)Energy metabolism Methanogenesis methyl coenzyme M reductase system, component A2 (TIGR03269; HMM-score: 95)ectoine/hydroxyectoine ABC transporter, ATP-binding protein EhuA (TIGR03005; HMM-score: 93.7)proposed F420-0 ABC transporter, ATP-binding protein (TIGR03873; HMM-score: 93.3)Transport and binding proteins Amino acids, peptides and amines choline ABC transporter, ATP-binding protein (TIGR03415; HMM-score: 92.8)gliding motility-associated ABC transporter ATP-binding subunit GldA (TIGR03522; HMM-score: 91.1)Cellular processes Other nodulation ABC transporter NodI (TIGR01288; HMM-score: 88.3)Transport and binding proteins Other nodulation ABC transporter NodI (TIGR01288; HMM-score: 88.3)Cellular processes Biosynthesis of natural products NHLM bacteriocin system ABC transporter, ATP-binding protein (TIGR03797; HMM-score: 86.3)Transport and binding proteins Amino acids, peptides and amines NHLM bacteriocin system ABC transporter, ATP-binding protein (TIGR03797; HMM-score: 86.3)Cellular processes Biosynthesis of natural products NHLM bacteriocin system ABC transporter, peptidase/ATP-binding protein (TIGR03796; HMM-score: 86.1)Transport and binding proteins Amino acids, peptides and amines NHLM bacteriocin system ABC transporter, peptidase/ATP-binding protein (TIGR03796; HMM-score: 86.1)Protein fate Protein and peptide secretion and trafficking type I secretion system ATPase (TIGR01842; HMM-score: 84.4)ABC transporter, permease/ATP-binding protein (TIGR02204; HMM-score: 83.3)Cell envelope Biosynthesis and degradation of surface polysaccharides and lipopolysaccharides lipid A export permease/ATP-binding protein MsbA (TIGR02203; HMM-score: 83.2)Transport and binding proteins Other lipid A export permease/ATP-binding protein MsbA (TIGR02203; HMM-score: 83.2)Cell envelope Biosynthesis and degradation of surface polysaccharides and lipopolysaccharides LPS export ABC transporter ATP-binding protein (TIGR04406; HMM-score: 82.5)Transport and binding proteins Other LPS export ABC transporter ATP-binding protein (TIGR04406; HMM-score: 82.5)Transport and binding proteins Amino acids, peptides and amines urea ABC transporter, ATP-binding protein UrtD (TIGR03411; HMM-score: 81.5)Protein fate Protein and peptide secretion and trafficking ABC-type bacteriocin transporter (TIGR01193; HMM-score: 81.4)Protein fate Protein modification and repair ABC-type bacteriocin transporter (TIGR01193; HMM-score: 81.4)Transport and binding proteins Other ABC-type bacteriocin transporter (TIGR01193; HMM-score: 81.4)Transport and binding proteins Cations and iron carrying compounds nickel import ATP-binding protein NikE (TIGR02769; EC 3.6.3.24; HMM-score: 80.4)lantibiotic protection ABC transporter, ATP-binding subunit (TIGR03740; HMM-score: 79.5)Transport and binding proteins Other antigen peptide transporter 2 (TIGR00958; HMM-score: 78.2)Protein fate Protein and peptide secretion and trafficking type I secretion system ATPase (TIGR01846; HMM-score: 76.4)Transport and binding proteins Cations and iron carrying compounds nickel import ATP-binding protein NikD (TIGR02770; HMM-score: 75.6)ATP-binding cassette protein, ChvD family (TIGR03719; HMM-score: 75.5)phosphonate C-P lyase system protein PhnL (TIGR02324; HMM-score: 64.3)Transport and binding proteins Carbohydrates, organic alcohols, and acids glucan exporter ATP-binding protein (TIGR01192; EC 3.6.3.42; HMM-score: 61)Biosynthesis of cofactors, prosthetic groups, and carriers Other FeS assembly ATPase SufC (TIGR01978; HMM-score: 60.7)Transport and binding proteins Unknown substrate anchored repeat-type ABC transporter, ATP-binding subunit (TIGR03771; HMM-score: 59.6)Transport and binding proteins Carbohydrates, organic alcohols, and acids D-xylose ABC transporter, ATP-binding protein (TIGR02633; EC 3.6.3.17; HMM-score: 53.9)Transport and binding proteins Other rim ABC transporter (TIGR01257; HMM-score: 51.5)DNA metabolism DNA replication, recombination, and repair excinuclease ABC subunit A (TIGR00630; EC 3.1.25.-; HMM-score: 51.3)Transport and binding proteins Other pleiotropic drug resistance family protein (TIGR00956; HMM-score: 49.9)Transport and binding proteins Amino acids, peptides and amines cyclic peptide transporter (TIGR01194; HMM-score: 49.7)Transport and binding proteins Other cyclic peptide transporter (TIGR01194; HMM-score: 49.7)Transport and binding proteins Other multi drug resistance-associated protein (MRP) (TIGR00957; HMM-score: 46.6)Transport and binding proteins Anions cystic fibrosis transmembrane conductor regulator (CFTR) (TIGR01271; EC 3.6.3.49; HMM-score: 46.4)Central intermediary metabolism Phosphorus compounds phosphonate C-P lyase system protein PhnK (TIGR02323; HMM-score: 42.7)Transport and binding proteins Carbohydrates, organic alcohols, and acids peroxysomal long chain fatty acyl transporter (TIGR00954; HMM-score: 39.3)Protein synthesis Translation factors ribosome small subunit-dependent GTPase A (TIGR00157; EC 3.6.-.-; HMM-score: 18.5)Protein synthesis Other ribosome biogenesis GTP-binding protein YsxC (TIGR03598; HMM-score: 16.2)Protein synthesis Other ribosome-associated GTPase EngA (TIGR03594; HMM-score: 15.7)Protein synthesis Other ribosome biogenesis GTPase YqeH (TIGR03597; HMM-score: 15.5)DNA metabolism DNA replication, recombination, and repair DnaA regulatory inactivator Hda (TIGR03420; HMM-score: 15.4)restriction system-associated AAA family ATPase (TIGR04435; HMM-score: 15.1)Unknown function General small GTP-binding protein domain (TIGR00231; HMM-score: 14.2)Energy metabolism Amino acids and amines ethanolamine utilization protein, EutP (TIGR02528; HMM-score: 13.8)DNA metabolism DNA replication, recombination, and repair exonuclease SbcC (TIGR00618; HMM-score: 10.9)
- TheSEED :
- Methionine ABC transporter ATP-binding protein
Amino Acids and Derivatives Lysine, threonine, methionine, and cysteine Methionine Biosynthesis Methionine ABC transporter ATP-binding proteinand 1 more - PFAM: P-loop_NTPase (CL0023) ABC_tran; ABC transporter (PF00005; HMM-score: 113.8)and 23 moreAAA_21; AAA domain, putative AbiEii toxin, Type IV TA system (PF13304; HMM-score: 26.7)RsgA_GTPase; RsgA GTPase (PF03193; HMM-score: 24.4)ATPase_2; ATPase domain predominantly from Archaea (PF01637; HMM-score: 23.4)AAA_15; AAA ATPase domain (PF13175; HMM-score: 19.8)SMC_N; RecF/RecN/SMC N terminal domain (PF02463; HMM-score: 19.6)AAA_29; P-loop containing region of AAA domain (PF13555; HMM-score: 19.5)AAA_14; AAA domain (PF13173; HMM-score: 18.5)MMR_HSR1; 50S ribosome-binding GTPase (PF01926; HMM-score: 18.1)NACHT; NACHT domain (PF05729; HMM-score: 17.3)DUF5906; Family of unknown function (DUF5906) (PF19263; HMM-score: 17.3)AAA_16; AAA ATPase domain (PF13191; HMM-score: 16.4)RNA_helicase; RNA helicase (PF00910; HMM-score: 15.4)FeoB_N; Ferrous iron transport protein B (PF02421; HMM-score: 15.2)AAA_22; AAA domain (PF13401; HMM-score: 15.2)G-alpha; G-protein alpha subunit (PF00503; HMM-score: 14.3)AAA_5; AAA domain (dynein-related subfamily) (PF07728; HMM-score: 14.3)Rad17; Rad17 P-loop domain (PF03215; HMM-score: 13.9)PduV-EutP; Ethanolamine utilisation - propanediol utilisation (PF10662; HMM-score: 13.2)DLIC; Dynein light intermediate chain (DLIC) (PF05783; HMM-score: 13.1)NPHP3_N; Nephrocystin 3, N-terminal (PF24883; HMM-score: 12.6)AAA_28; AAA domain (PF13521; HMM-score: 12.2)nSTAND1; Novel STAND NTPase 1 (PF20703; HMM-score: 12.1)ATP-synt_ab; ATP synthase alpha/beta family, nucleotide-binding domain (PF00006; HMM-score: 11.6)
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors:
- effectors:
⊟Localization[edit | edit source]
- PSORTb: Cytoplasmic Membrane
- Cytoplasmic Score: 0.01
- Cytoplasmic Membrane Score: 9.99
- Cellwall Score: 0
- Extracellular Score: 0
- Internal Helices: 0
- DeepLocPro: Cytoplasmic Membrane
- Cytoplasmic Score: 0.4078
- Cytoplasmic Membrane Score: 0.558
- Cell wall & surface Score: 0.0007
- Extracellular Score: 0.0335
- LocateP: Intracellular
- Prediction by SwissProt Classification: Cytoplasmic
- Pathway Prediction: No pathway
- Intracellular possibility: 1
- Signal peptide possibility: -1
- N-terminally Anchored Score: -1
- Predicted Cleavage Site: No CleavageSite
- SignalP: no predicted signal peptide
- SP(Sec/SPI): 0.139368
- TAT(Tat/SPI): 0.002686
- LIPO(Sec/SPII): 0.056803
- predicted transmembrane helices (TMHMM): 0
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MIELKDLTIQKGNTHILKKLNLKFQCGKSYALIGKSGCGKSTLLNTIAGLEKTGKQYVYFNGQLEQFKSNFYRDKLGYLFQNYGLIDNLTVNENLDIGLAYKKISKKEKEQIKIRYIEQFGLSNSLKRKVHTLSGGEQQRVALIRMMLKDPIVMLADEPTGALDPKTGQMIIQSLFGLVDENKVLILATHDMAIANQCDEIIDLEQYRKVASM
⊟Experimental data[edit | edit source]
- experimentally validated:
- protein localization: data available for COL
- quantitative data / protein copy number per cell:
- interaction partners:
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
⊟Regulation[edit | edit source]
- regulator:
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: [1]
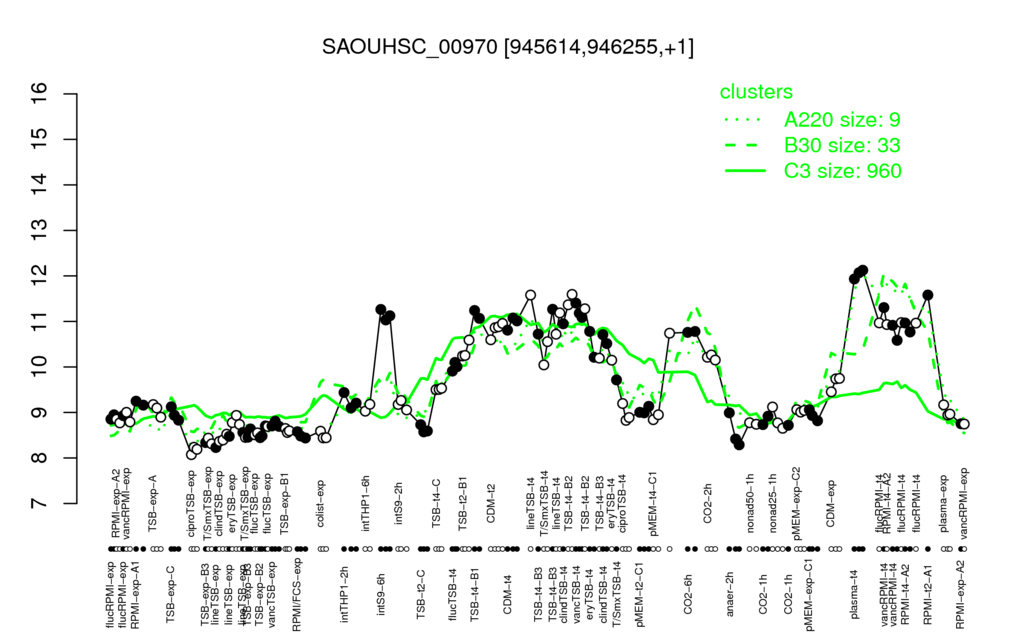 Multi-gene expression profiles
Multi-gene expression profiles
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
- Aureolib: no data available
⊟Protein stability[edit | edit source]
- half-life: no data available
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You can add further information about the gene and protein here. [edit]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ 1.0 1.1 Ulrike Mäder, Pierre Nicolas, Maren Depke, Jan Pané-Farré, Michel Debarbouille, Magdalena M van der Kooi-Pol, Cyprien Guérin, Sandra Dérozier, Aurelia Hiron, Hanne Jarmer, Aurélie Leduc, Stephan Michalik, Ewoud Reilman, Marc Schaffer, Frank Schmidt, Philippe Bessières, Philippe Noirot, Michael Hecker, Tarek Msadek, Uwe Völker, Jan Maarten van Dijl
Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions.
PLoS Genet: 2016, 12(4);e1005962
[PubMed:27035918] [WorldCat.org] [DOI] (I e)
