Jump to navigation
Jump to search
PangenomeCOLN315NCTC8325NewmanUSA300_FPR3757JSNZ04-0298108BA0217611819-97685071193ECT-R 2ED133ED98HO 5096 0412JH1JH9JKD6008JKD6159LGA251M013MRSA252MSHR1132MSSA476MW2Mu3Mu50RF122ST398T0131TCH60TW20USA300_TCH1516VC40
NCBI: 03-AUG-2016
⊟Summary[edit | edit source]
- organism: Staphylococcus aureus NCTC8325
- locus tag: SAOUHSC_02472
- pan locus tag?: SAUPAN005646000
- symbol: SAOUHSC_02472
- pan gene symbol?: —
- synonym:
- product: hypothetical protein
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
- type: CDS
- locus tag: SAOUHSC_02472
- symbol: SAOUHSC_02472
- product: hypothetical protein
- replicon: chromosome
- strand: -
- coordinates: 2295453..2296589
- length: 1137
- essential: no DEG
⊟Accession numbers[edit | edit source]
- Gene ID: 3919035 NCBI
- RefSeq: YP_500940 NCBI
- BioCyc: G1I0R-2337 BioCyc
- MicrobesOnline: 1290911 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421
481
541
601
661
721
781
841
901
961
1021
1081ATGGAAATTCATGTTTATACGGGACCGCTTTGTGGATTTAATGAATATATTAATTCTGAA
GTTAAAGAATATCACCGCTTCATGGAATTAATCAGAGAATACAATATCATAGTGCGAGCA
AGTGATACATTTGGAACTCATGATGTTGACGGTATTTTTGATACTGAGATAGAAAATCTT
GTGATATTTTCAAACGACTTTGCTTCTGTAACAACTCATGTTATTACTAATTTTATAAAT
ATCGTTACTTTAGGTAGAAATATTAAGAAAATCTATATTCAGAATCCTCCAAAAAGAGTA
TTTCAATCACTAAATAGTTTTCACGATTCTGAAATAATAGAAATAAACTATGAATATACA
ACATTAAAAAAAGATGATGTAATAAATTTTTATAAAGAGCAGATGGCTGGTTCTAAAATA
ATAGGTCAAACAGACGCTAAAAAACAAATGAGCATAGATTTATATAAACATACTTCCTTA
GAAAATAATAAACCGACCGTAATGCTTTACTATGGTCCATCAGGTGTTGGAAAAACAGAA
TTAGCCAAAGAAATAGCAAAAAAATATAAAGGTAATATCACTAGAATACAATTTTCTATG
ATGCAGAATGATGAATCTTTTAATTATATCTTTGGAGATGAACATGCCAAAGTATCATTG
TCAAAAGATTTAGTAGATCGTGAAACTAATGTTGTTTTAATTGATGAATTTGATAAGGTA
TTACCAATCTATTACAATGTTTTCTATCAAATGTTTGATGAAGGTATTATTGAAGATGCC
AATTATAAAGTAGACGTTTCAAATTGTATTTTCATTCTAACATCTAATTTTAATAGTATT
GAAGAAGCAATAGTTAATATTGGAGAACCGGTATTTTCTAGAATAGATTCCTGTATTAAA
TTTTTATATTTAACACAGGAAGAAAAGCAGAAAGTAATAGAAAATAAATTAAATGAAATA
CTATCATCTCTAAATGAAAAAGAAAAAAGAATAGTTACCGCATATAACTTGTTAGAGAAA
TATAAAGAGACAGAAATAGATATCGATAACATTAGGTATTTAAAGAAAATAATAACAAAT
GATATTTATACAATAATTTTTGAAGATGAATTATTTAATACGGATATTATAGATTAA60
120
180
240
300
360
420
480
540
600
660
720
780
840
900
960
1020
1080
1137
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SAOUHSC_02472
- symbol: SAOUHSC_02472
- description: hypothetical protein
- length: 378
- theoretical pI: 4.58881
- theoretical MW: 44099.8
- GRAVY: -0.289947
⊟Function[edit | edit source]
- TIGRFAM: Protein fate Degradation of proteins, peptides, and glycopeptides ATP-dependent Clp protease ATP-binding subunit ClpA (TIGR02639; HMM-score: 67.3)Cellular processes Pathogenesis type VI secretion ATPase, ClpV1 family (TIGR03345; HMM-score: 61.1)Protein fate Protein and peptide secretion and trafficking type VI secretion ATPase, ClpV1 family (TIGR03345; HMM-score: 61.1)Protein fate Protein folding and stabilization ATP-dependent chaperone protein ClpB (TIGR03346; HMM-score: 57)and 12 moreDNA metabolism DNA replication, recombination, and repair Holliday junction DNA helicase RuvB (TIGR00635; EC 3.6.4.12; HMM-score: 33.1)Protein fate Degradation of proteins, peptides, and glycopeptides endopeptidase La (TIGR00763; EC 3.4.21.53; HMM-score: 24.8)Protein fate Protein folding and stabilization ATP-dependent protease HslVU, ATPase subunit (TIGR00390; HMM-score: 23.6)Cellular processes Other gas vesicle protein GvpN (TIGR02640; HMM-score: 19.1)26S proteasome subunit P45 family (TIGR01242; HMM-score: 18.3)AAA family ATPase, CDC48 subfamily (TIGR01243; HMM-score: 17.2)DNA metabolism DNA replication, recombination, and repair orc1/cdc6 family replication initiation protein (TIGR02928; HMM-score: 15.6)Protein fate Degradation of proteins, peptides, and glycopeptides proteasome ATPase (TIGR03689; EC 3.6.4.8; HMM-score: 15)Cellular processes Chemotaxis and motility flagellar biosynthesis protein FlhF (TIGR03499; HMM-score: 13.4)Protein fate Protein and peptide secretion and trafficking signal recognition particle-docking protein FtsY (TIGR00064; HMM-score: 12.8)phosphonate C-P lyase system protein PhnL (TIGR02324; HMM-score: 12.5)Protein fate Protein and peptide secretion and trafficking type VII secretion AAA-ATPase EccA (TIGR03922; HMM-score: 11)
- TheSEED :
- ClpB protein
and 1 more - PFAM: P-loop_NTPase (CL0023) AAA_2; AAA domain (Cdc48 subfamily) (PF07724; HMM-score: 66.4)and 26 moreAAA; ATPase family associated with various cellular activities (AAA) (PF00004; HMM-score: 43.9)AAA_5; AAA domain (dynein-related subfamily) (PF07728; HMM-score: 28.1)AAA_3; ATPase family associated with various cellular activities (AAA) (PF07726; HMM-score: 24.3)RuvB_N; Holliday junction DNA helicase RuvB P-loop domain (PF05496; HMM-score: 23.6)AAA_14; AAA domain (PF13173; HMM-score: 22)nSTAND3; Novel STAND NTPase 3 (PF20720; HMM-score: 20.2)RNA_helicase; RNA helicase (PF00910; HMM-score: 19.4)AAA_22; AAA domain (PF13401; HMM-score: 19.4)NB-ARC; NB-ARC domain (PF00931; HMM-score: 19)AAA_16; AAA ATPase domain (PF13191; HMM-score: 18.4)ATPase_2; ATPase domain predominantly from Archaea (PF01637; HMM-score: 16.2)Rad17; Rad17 P-loop domain (PF03215; HMM-score: 15.9)ABC_tran; ABC transporter (PF00005; HMM-score: 15.8)AAA_17; AAA domain (PF13207; HMM-score: 15.5)Bac_DnaA; Bacterial DnaA ATPAse domain (PF00308; HMM-score: 15.4)TsaE; Threonylcarbamoyl adenosine biosynthesis protein TsaE (PF02367; HMM-score: 15.3)AAA_18; AAA domain (PF13238; HMM-score: 15.3)AAA_7; P-loop containing dynein motor region (PF12775; HMM-score: 15.2)AAA_24; AAA domain (PF13479; HMM-score: 15)Zeta_toxin; Zeta toxin (PF06414; HMM-score: 14.4)NPHP3_N; Nephrocystin 3, N-terminal (PF24883; HMM-score: 13.6)RsgA_GTPase; RsgA GTPase (PF03193; HMM-score: 12.9)AAA_33; AAA domain (PF13671; HMM-score: 12.6)IstB_IS21; IstB-like ATP binding protein (PF01695; HMM-score: 12)NACHT; NACHT domain (PF05729; HMM-score: 11.9)AAA_25; AAA domain (PF13481; HMM-score: 11.1)
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors:
- effectors:
⊟Localization[edit | edit source]
- PSORTb: Cytoplasmic
- Cytoplasmic Score: 9.97
- Cytoplasmic Membrane Score: 0
- Cellwall Score: 0.01
- Extracellular Score: 0.02
- Internal Helices: 0
- DeepLocPro: Cytoplasmic
- Cytoplasmic Score: 0.572
- Cytoplasmic Membrane Score: 0.1731
- Cell wall & surface Score: 0.0016
- Extracellular Score: 0.2532
- LocateP: Intracellular
- Prediction by SwissProt Classification: Cytoplasmic
- Pathway Prediction: No pathway
- Intracellular possibility: 1
- Signal peptide possibility: -1
- N-terminally Anchored Score: 1
- Predicted Cleavage Site: No CleavageSite
- SignalP: no predicted signal peptide
- SP(Sec/SPI): 0.003073
- TAT(Tat/SPI): 0.000098
- LIPO(Sec/SPII): 0.000419
- predicted transmembrane helices (TMHMM): 0
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MEIHVYTGPLCGFNEYINSEVKEYHRFMELIREYNIIVRASDTFGTHDVDGIFDTEIENLVIFSNDFASVTTHVITNFINIVTLGRNIKKIYIQNPPKRVFQSLNSFHDSEIIEINYEYTTLKKDDVINFYKEQMAGSKIIGQTDAKKQMSIDLYKHTSLENNKPTVMLYYGPSGVGKTELAKEIAKKYKGNITRIQFSMMQNDESFNYIFGDEHAKVSLSKDLVDRETNVVLIDEFDKVLPIYYNVFYQMFDEGIIEDANYKVDVSNCIFILTSNFNSIEEAIVNIGEPVFSRIDSCIKFLYLTQEEKQKVIENKLNEILSSLNEKEKRIVTAYNLLEKYKETEIDIDNIRYLKKIITNDIYTIIFEDELFNTDIID
⊟Experimental data[edit | edit source]
- experimentally validated: PeptideAtlas [1] [2]
- protein localization: data available for COL
- quantitative data / protein copy number per cell:
- interaction partners:
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
- predicted SigA promoter [3] : SAOUHSC_02470 < S960 < SAOUHSC_02471 < S961 < SAOUHSC_02472 < SAOUHSC_02474
⊟Regulation[edit | edit source]
- regulator:
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: [3]
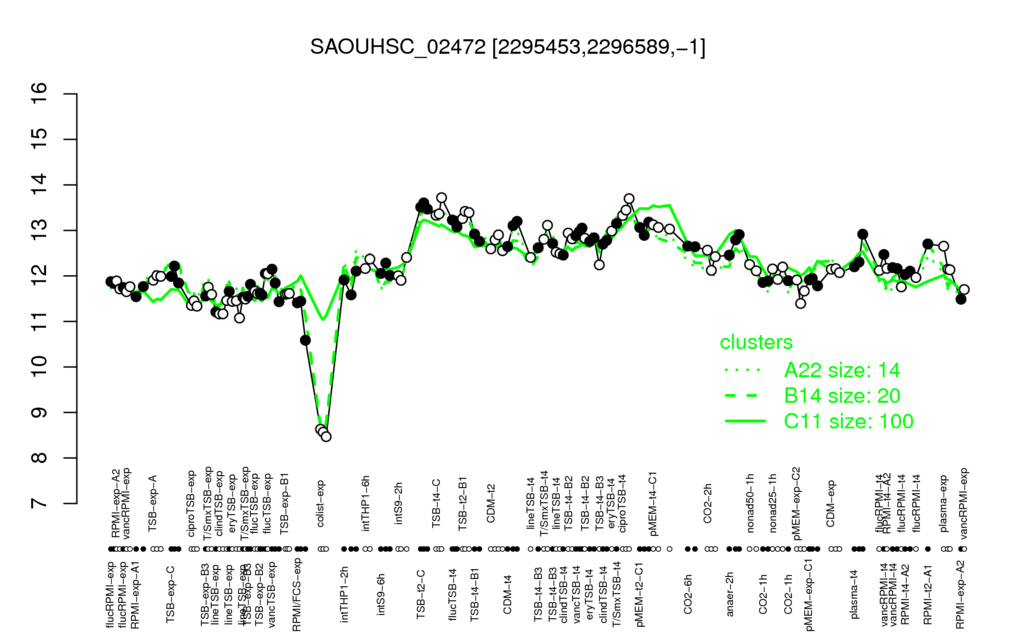 Multi-gene expression profiles
Multi-gene expression profiles
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
- Aureolib: no data available
⊟Protein stability[edit | edit source]
- half-life: no data available
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You can add further information about the gene and protein here. [edit]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ Maren Depke, Stephan Michalik, Alexander Rabe, Kristin Surmann, Lars Brinkmann, Nico Jehmlich, Jörg Bernhardt, Michael Hecker, Bernd Wollscheid, Zhi Sun, Robert L Moritz, Uwe Völker, Frank Schmidt
A peptide resource for the analysis of Staphylococcus aureus in host-pathogen interaction studies.
Proteomics: 2015, 15(21);3648-61
[PubMed:26224020] [WorldCat.org] [DOI] (I p) - ↑ Stephan Michalik, Maren Depke, Annette Murr, Manuela Gesell Salazar, Ulrike Kusebauch, Zhi Sun, Tanja C Meyer, Kristin Surmann, Henrike Pförtner, Petra Hildebrandt, Stefan Weiss, Laura Marcela Palma Medina, Melanie Gutjahr, Elke Hammer, Dörte Becher, Thomas Pribyl, Sven Hammerschmidt, Eric W Deutsch, Samuel L Bader, Michael Hecker, Robert L Moritz, Ulrike Mäder, Uwe Völker, Frank Schmidt
A global Staphylococcus aureus proteome resource applied to the in vivo characterization of host-pathogen interactions.
Sci Rep: 2017, 7(1);9718
[PubMed:28887440] [WorldCat.org] [DOI] (I e) - ↑ 3.0 3.1 Ulrike Mäder, Pierre Nicolas, Maren Depke, Jan Pané-Farré, Michel Debarbouille, Magdalena M van der Kooi-Pol, Cyprien Guérin, Sandra Dérozier, Aurelia Hiron, Hanne Jarmer, Aurélie Leduc, Stephan Michalik, Ewoud Reilman, Marc Schaffer, Frank Schmidt, Philippe Bessières, Philippe Noirot, Michael Hecker, Tarek Msadek, Uwe Völker, Jan Maarten van Dijl
Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions.
PLoS Genet: 2016, 12(4);e1005962
[PubMed:27035918] [WorldCat.org] [DOI] (I e)
