Jump to navigation
Jump to search
NCBI: 03-AUG-2016
⊟Summary[edit | edit source]
- organism: Staphylococcus aureus NCTC8325
- locus tag: SAOUHSC_00242
- pan locus tag?: SAUPAN001157000
- symbol: SAOUHSC_00242
- pan gene symbol?: rbsR
- synonym:
- product: hypothetical protein
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
- type: CDS
- locus tag: SAOUHSC_00242
- symbol: SAOUHSC_00242
- product: hypothetical protein
- replicon: chromosome
- strand: -
- coordinates: 262382..263380
- length: 999
- essential: no DEG other strains
⊟Accession numbers[edit | edit source]
- Gene ID: 3919262 NCBI
- RefSeq: YP_498837 NCBI
- BioCyc: G1I0R-226 BioCyc
- MicrobesOnline: 1288731 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421
481
541
601
661
721
781
841
901
961ATGAAAAAAGTGTCAATTAAAGATGTTGCTAGAGAAGCTGGTGTATCAGTTACAACTGTG
TCACATATTTTAAATCATAATGATAGTCGTTTTTCCGCAACAACGATAAAAAACGTACAT
GCTGTTTCAGAACGTTTAGGCTATGCCCCTAATAAACATGCAAAACAATTGCGCGGCAGT
AAAATTCAAACTATTGGCGTCATTTTGCCTAGCTTAACAAATCCGTTTTTCTCAGCACTG
ATGCAAAGTATTCATGACCATAAACCATCTGATGTTGATTTATGCTTTTTAACATCTACA
GCAACTGATTTGTATGACAATATTAAACATTTAATTGATCGAGGTATTGACGGATTAATT
ATCGCACAATACATATCATCCCCGGACGCCCTAAATAACTATCTAAAGAAACATCATGTA
CCTTATGTCGTACTGGATCAAAATGACCATCAAGGCTATACAGATTTTGTTCGGACAAAT
GAATATCAAGGTGGACAACTTGCAGCACAACATTTAATAGAACTCGGTCACAACCATATG
ATAATTGTTGCACCATATGACATGATGGCGAATATGTCGACTCGTGTCGCTGGATTTGTC
GATACTTTGCGCGCGAATCAATTGCCAGAACCACAAATCGTCCATACTGAATTATCTAAG
CGCGGTGGGCTAACCATTGTTGATGACATCATGGTTCAATCTGCCACTGCAATCTTCGCT
ATTAACGATGAACTCGCTATTGGCATTTTACGAGGACTAATTGAACATGGCATCAGTATC
CCGAAAGATATCTCATTAATAGGTTATGACGACATTGATTATGCAGCGTACGTCTCGCCA
CCTTTAACTACTGTGGCACAACCTATAACTGATATTGGCAAAACATCTTTAACCTTATTA
CTTCAACGATTACAGCACTTAGATAAATCCATTGATATGATTGAATTACCAACGACTTTA
AAAATTCGTGCAACAACTGGCTATCATCTTTCAAACTAA60
120
180
240
300
360
420
480
540
600
660
720
780
840
900
960
999
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SAOUHSC_00242
- symbol: SAOUHSC_00242
- description: hypothetical protein
- length: 332
- theoretical pI: 6.76486
- theoretical MW: 36657.7
- GRAVY: -0.0650602
⊟Function[edit | edit source]
- TIGRFAM: Regulatory functions DNA interactions catabolite control protein A (TIGR01481; HMM-score: 172.7)and 4 moreRegulatory functions DNA interactions D-fructose-responsive transcription factor (TIGR02417; HMM-score: 117.9)Regulatory functions DNA interactions trehalose operon repressor (TIGR02405; HMM-score: 74.3)Regulatory functions DNA interactions transcriptional repressor BetI (TIGR03384; HMM-score: 13.9)Protein synthesis tRNA aminoacylation aspartyl/glutamyl-tRNA(Asn/Gln) amidotransferase, A subunit (TIGR00132; EC 6.3.5.-; HMM-score: 11)
- TheSEED :
- Ribose operon repressor RbsR, LacI family
- PFAM: Periplas_BP (CL0144) Peripla_BP_3; Periplasmic binding protein-like domain (PF13377; HMM-score: 116.5)and 17 morePeripla_BP_1; Periplasmic binding proteins and sugar binding domain of LacI family (PF00532; HMM-score: 83.6)Peripla_BP_4; Periplasmic binding protein domain (PF13407; HMM-score: 58.5)HTH (CL0123) LacI; Bacterial regulatory proteins, lacI family (PF00356; HMM-score: 55.3)HTH_3; Helix-turn-helix (PF01381; HMM-score: 18.6)HTH_Tnp_ISL3; Helix-turn-helix domain of transposase family ISL3 (PF13542; HMM-score: 18.1)HTH_31; Helix-turn-helix domain (PF13560; HMM-score: 16.4)HTH_26; Cro/C1-type HTH DNA-binding domain (PF13443; HMM-score: 16)HTH_19; Helix-turn-helix domain (PF12844; HMM-score: 13.9)HTH_7; Helix-turn-helix domain of resolvase (PF02796; HMM-score: 13.8)TetR_N; Bacterial regulatory proteins, tetR family (PF00440; HMM-score: 13.5)HTH_38; Helix-turn-helix domain (PF13936; HMM-score: 13.4)Gain (CL0661) GPR128_GAIN_subdomA; GPR128, GAIN subdomain A (PF22259; HMM-score: 13)HTH (CL0123) HTH_10; HTH DNA binding domain (PF04967; HMM-score: 12.7)no clan defined PucR; Purine catabolism regulatory protein-like family (PF07905; HMM-score: 12.5)HTH (CL0123) HTH_23; Homeodomain-like domain (PF13384; HMM-score: 12)MerR; MerR family regulatory protein (PF00376; HMM-score: 11.9)TnsD; Tn7-like transposition protein D (PF15978; HMM-score: 10.7)
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors:
- effector: Ribose
⊟Localization[edit | edit source]
- PSORTb: Cytoplasmic
- Cytoplasmic Score: 9.67
- Cytoplasmic Membrane Score: 0.01
- Cellwall Score: 0.15
- Extracellular Score: 0.17
- Internal Helices: 0
- DeepLocPro: Cytoplasmic
- Cytoplasmic Score: 0.9922
- Cytoplasmic Membrane Score: 0.0022
- Cell wall & surface Score: 0.0001
- Extracellular Score: 0.0055
- LocateP: Intracellular
- Prediction by SwissProt Classification: Cytoplasmic
- Pathway Prediction: No pathway
- Intracellular possibility: 1
- Signal peptide possibility: -1
- N-terminally Anchored Score: -1
- Predicted Cleavage Site: No CleavageSite
- SignalP: no predicted signal peptide
- SP(Sec/SPI): 0.026708
- TAT(Tat/SPI): 0.005312
- LIPO(Sec/SPII): 0.001669
- predicted transmembrane helices (TMHMM): 0
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MKKVSIKDVAREAGVSVTTVSHILNHNDSRFSATTIKNVHAVSERLGYAPNKHAKQLRGSKIQTIGVILPSLTNPFFSALMQSIHDHKPSDVDLCFLTSTATDLYDNIKHLIDRGIDGLIIAQYISSPDALNNYLKKHHVPYVVLDQNDHQGYTDFVRTNEYQGGQLAAQHLIELGHNHMIIVAPYDMMANMSTRVAGFVDTLRANQLPEPQIVHTELSKRGGLTIVDDIMVQSATAIFAINDELAIGILRGLIEHGISIPKDISLIGYDDIDYAAYVSPPLTTVAQPITDIGKTSLTLLLQRLQHLDKSIDMIELPTTLKIRATTGYHLSN
⊟Experimental data[edit | edit source]
- experimentally validated: PeptideAtlas [2] [3]
- protein localization: data available for COL
- quantitative data / protein copy number per cell:
- interaction partners:
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
- MicrobesOnline: no polycistronic organisation predicted
⊟Regulation[edit | edit source]
- regulator: SigB* (activation) regulon
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: [1]
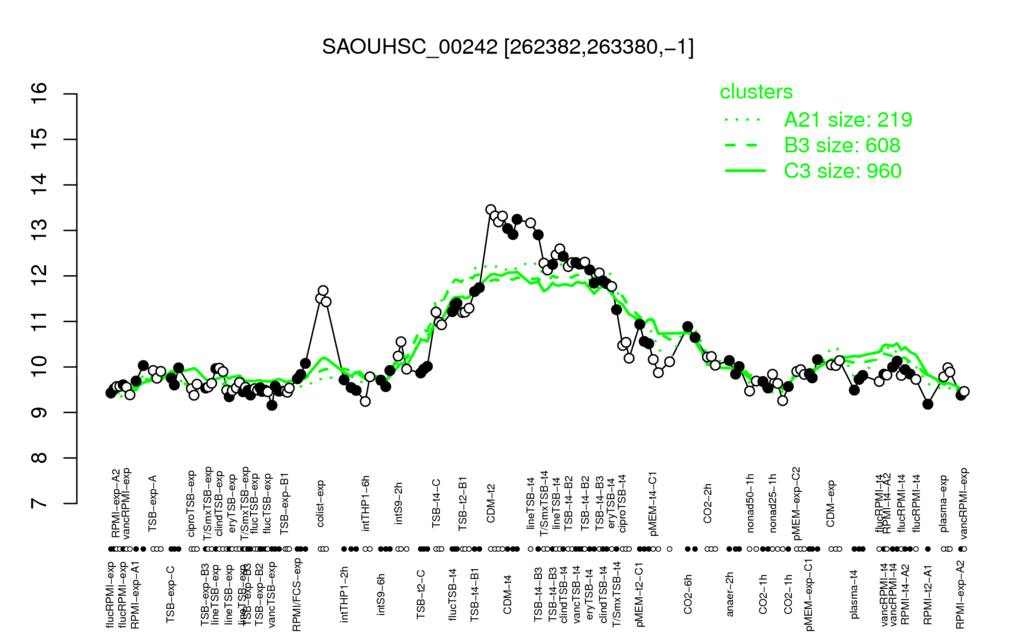 Multi-gene expression profiles
Multi-gene expression profiles
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
⊟Protein stability[edit | edit source]
- half-life: no data available
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You can add further information about the gene and protein here. [edit]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ 1.0 1.1 1.2 Ulrike Mäder, Pierre Nicolas, Maren Depke, Jan Pané-Farré, Michel Debarbouille, Magdalena M van der Kooi-Pol, Cyprien Guérin, Sandra Dérozier, Aurelia Hiron, Hanne Jarmer, Aurélie Leduc, Stephan Michalik, Ewoud Reilman, Marc Schaffer, Frank Schmidt, Philippe Bessières, Philippe Noirot, Michael Hecker, Tarek Msadek, Uwe Völker, Jan Maarten van Dijl
Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions.
PLoS Genet: 2016, 12(4);e1005962
[PubMed:27035918] [WorldCat.org] [DOI] (I e) - ↑ Maren Depke, Stephan Michalik, Alexander Rabe, Kristin Surmann, Lars Brinkmann, Nico Jehmlich, Jörg Bernhardt, Michael Hecker, Bernd Wollscheid, Zhi Sun, Robert L Moritz, Uwe Völker, Frank Schmidt
A peptide resource for the analysis of Staphylococcus aureus in host-pathogen interaction studies.
Proteomics: 2015, 15(21);3648-61
[PubMed:26224020] [WorldCat.org] [DOI] (I p) - ↑ Stephan Michalik, Maren Depke, Annette Murr, Manuela Gesell Salazar, Ulrike Kusebauch, Zhi Sun, Tanja C Meyer, Kristin Surmann, Henrike Pförtner, Petra Hildebrandt, Stefan Weiss, Laura Marcela Palma Medina, Melanie Gutjahr, Elke Hammer, Dörte Becher, Thomas Pribyl, Sven Hammerschmidt, Eric W Deutsch, Samuel L Bader, Michael Hecker, Robert L Moritz, Ulrike Mäder, Uwe Völker, Frank Schmidt
A global Staphylococcus aureus proteome resource applied to the in vivo characterization of host-pathogen interactions.
Sci Rep: 2017, 7(1);9718
[PubMed:28887440] [WorldCat.org] [DOI] (I e) - ↑ Markus Bischoff, Paul Dunman, Jan Kormanec, Daphne Macapagal, Ellen Murphy, William Mounts, Brigitte Berger-Bächi, Steven Projan
Microarray-based analysis of the Staphylococcus aureus sigmaB regulon.
J Bacteriol: 2004, 186(13);4085-99
[PubMed:15205410] [WorldCat.org] [DOI] (P p)
