⊟Summary[edit | edit source]
- organism: Staphylococcus aureus NCTC8325
- locus tag: SAOUHSC_00620
- pan locus tag?: SAUPAN002485000
- symbol: SAOUHSC_00620
- pan gene symbol?: sarA
- synonym:
- product: accessory regulator A
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
⊟Accession numbers[edit | edit source]
- Gene ID: 3918939 NCBI
- RefSeq: YP_499183 NCBI
- BioCyc: G1I0R-580 BioCyc
- MicrobesOnline: 1289093 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361ATGGCAATTACAAAAATCAATGATTGCTTTGAGTTGTTATCAATGGTCACTTATGCTGAC
AAATTAAAAAGTTTAATTAAAAAGGAATTTTCAATTAGCTTTGAAGAATTCGCTGTATTG
ACATACATCAGCGAAAACAAAGAGAAAGAATACTATCTTAAAGATATTATTAATCATTTA
AACTACAAACAACCACAAGTTGTTAAAGCAGTTAAAATTTTATCTCAAGAAGATTACTTC
GATAAAAAACGTAATGAGCATGATGAAAGAACTGTATTAATTCTTGTTAATGCACAACAA
CGTAAAAAAATCGAATCATTATTGAGTCGAGTAAATAAACGAATCACTGAAGCAAACAAC
GAAATTGAACTATAA60
120
180
240
300
360
375
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SAOUHSC_00620
- symbol: SAOUHSC_00620
- description: accessory regulator A
- length: 124
- theoretical pI: 8.4094
- theoretical MW: 14717.9
- GRAVY: -0.495968
⊟Function[edit | edit source]
- TIGRFAM: Regulatory functions DNA interactions staphylococcal accessory regulator family (TIGR01889; HMM-score: 118)and 6 moreCell envelope Biosynthesis and degradation of surface polysaccharides and lipopolysaccharides UDP-N-acetylglucosamine diphosphorylase/glucosamine-1-phosphate N-acetyltransferase (TIGR01173; EC 2.3.1.157,2.7.7.23; HMM-score: 13.3)Cell envelope Biosynthesis and degradation of murein sacculus and peptidoglycan UDP-N-acetylglucosamine diphosphorylase/glucosamine-1-phosphate N-acetyltransferase (TIGR01173; EC 2.3.1.157,2.7.7.23; HMM-score: 13.3)Central intermediary metabolism Amino sugars UDP-N-acetylglucosamine diphosphorylase/glucosamine-1-phosphate N-acetyltransferase (TIGR01173; EC 2.3.1.157,2.7.7.23; HMM-score: 13.3)homoprotocatechuate degradation operon regulator, HpaR (TIGR02337; HMM-score: 13)undecaprenyl-phosphate glucose phosphotransferase (TIGR03023; EC 2.7.8.-; HMM-score: 12.2)Protein fate Protein modification and repair [FeFe] hydrogenase H-cluster maturation GTPase HydF (TIGR03918; HMM-score: 11.3)
- TheSEED :
- Staphylococcal accessory regulator A (SarA)
- PFAM: HTH (CL0123) Staph_reg_Sar_Rot; Transcriptional regulator SarA/Rot (PF22381; HMM-score: 87.2)and 4 moreHTH_27; Winged helix DNA-binding domain (PF13463; HMM-score: 25)MarR_2; MarR family (PF12802; HMM-score: 22.4)MarR; MarR family (PF01047; HMM-score: 19.9)P-loop_NTPase (CL0023) AAA_23; AAA domain (PF13476; HMM-score: 11.7)
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors:
- effectors:
- genes regulated by SarA, TF important in Virulence: in JSNZ
⊟Localization[edit | edit source]
- PSORTb: Cytoplasmic
- Cytoplasmic Score: 10
- Cytoplasmic Membrane Score: 0
- Cellwall Score: 0
- Extracellular Score: 0
- Internal Helices: 0
- DeepLocPro: Cytoplasmic
- Cytoplasmic Score: 0.9019
- Cytoplasmic Membrane Score: 0.0091
- Cell wall & surface Score: 0.0007
- Extracellular Score: 0.0884
- LocateP: Intracellular
- Prediction by SwissProt Classification: Cytoplasmic
- Pathway Prediction: No pathway
- Intracellular possibility: 1
- Signal peptide possibility: -1
- N-terminally Anchored Score: -1
- Predicted Cleavage Site: No CleavageSite
- SignalP: no predicted signal peptide
- SP(Sec/SPI): 0.004617
- TAT(Tat/SPI): 0.000227
- LIPO(Sec/SPII): 0.000605
- predicted transmembrane helices (TMHMM): 0
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MAITKINDCFELLSMVTYADKLKSLIKKEFSISFEEFAVLTYISENKEKEYYLKDIINHLNYKQPQVVKAVKILSQEDYFDKKRNEHDERTVLILVNAQQRKKIESLLSRVNKRITEANNEIEL
⊟Experimental data[edit | edit source]
- experimentally validated: PeptideAtlas [2] [3]
- protein localization: data available for COL
- quantitative data / protein copy number per cell: data available for COL
- interaction partners:
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
- predicted SigA promoter [4] : S226 < SAOUHSC_00620 < S227 < S228
⊟Regulation[edit | edit source]
- regulator: SigB* (activation) regulon
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: [4]
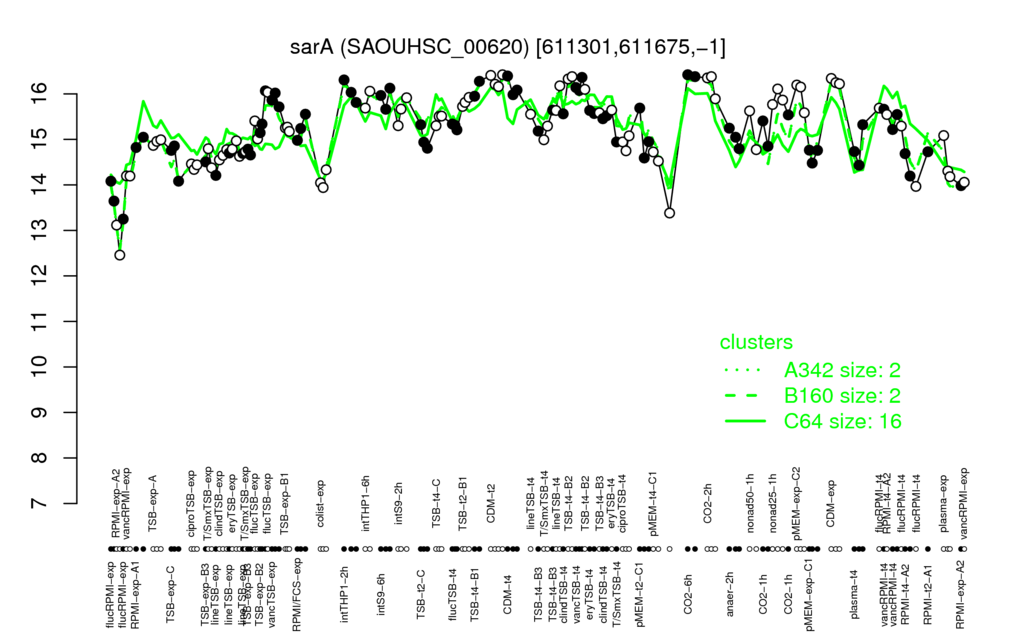 Multi-gene expression profiles
Multi-gene expression profiles
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
- Aureolib: no data available
⊟Protein stability[edit | edit source]
- half-life: no data available
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You can add further information about the gene and protein here. [edit]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ Roy R Chaudhuri, Andrew G Allen, Paul J Owen, Gil Shalom, Karl Stone, Marcus Harrison, Timothy A Burgis, Michael Lockyer, Jorge Garcia-Lara, Simon J Foster, Stephen J Pleasance, Sarah E Peters, Duncan J Maskell, Ian G Charles
Comprehensive identification of essential Staphylococcus aureus genes using Transposon-Mediated Differential Hybridisation (TMDH).
BMC Genomics: 2009, 10;291
[PubMed:19570206] [WorldCat.org] [DOI] (I e) - ↑ Maren Depke, Stephan Michalik, Alexander Rabe, Kristin Surmann, Lars Brinkmann, Nico Jehmlich, Jörg Bernhardt, Michael Hecker, Bernd Wollscheid, Zhi Sun, Robert L Moritz, Uwe Völker, Frank Schmidt
A peptide resource for the analysis of Staphylococcus aureus in host-pathogen interaction studies.
Proteomics: 2015, 15(21);3648-61
[PubMed:26224020] [WorldCat.org] [DOI] (I p) - ↑ Stephan Michalik, Maren Depke, Annette Murr, Manuela Gesell Salazar, Ulrike Kusebauch, Zhi Sun, Tanja C Meyer, Kristin Surmann, Henrike Pförtner, Petra Hildebrandt, Stefan Weiss, Laura Marcela Palma Medina, Melanie Gutjahr, Elke Hammer, Dörte Becher, Thomas Pribyl, Sven Hammerschmidt, Eric W Deutsch, Samuel L Bader, Michael Hecker, Robert L Moritz, Ulrike Mäder, Uwe Völker, Frank Schmidt
A global Staphylococcus aureus proteome resource applied to the in vivo characterization of host-pathogen interactions.
Sci Rep: 2017, 7(1);9718
[PubMed:28887440] [WorldCat.org] [DOI] (I e) - ↑ 4.0 4.1 4.2 4.3 Ulrike Mäder, Pierre Nicolas, Maren Depke, Jan Pané-Farré, Michel Debarbouille, Magdalena M van der Kooi-Pol, Cyprien Guérin, Sandra Dérozier, Aurelia Hiron, Hanne Jarmer, Aurélie Leduc, Stephan Michalik, Ewoud Reilman, Marc Schaffer, Frank Schmidt, Philippe Bessières, Philippe Noirot, Michael Hecker, Tarek Msadek, Uwe Völker, Jan Maarten van Dijl
Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions.
PLoS Genet: 2016, 12(4);e1005962
[PubMed:27035918] [WorldCat.org] [DOI] (I e) - ↑ Markus Bischoff, Paul Dunman, Jan Kormanec, Daphne Macapagal, Ellen Murphy, William Mounts, Brigitte Berger-Bächi, Steven Projan
Microarray-based analysis of the Staphylococcus aureus sigmaB regulon.
J Bacteriol: 2004, 186(13);4085-99
[PubMed:15205410] [WorldCat.org] [DOI] (P p)
⊟Relevant publications[edit | edit source]
J S Blevins, A F Gillaspy, T M Rechtin, B K Hurlburt, M S Smeltzer
The Staphylococcal accessory regulator (sar) represses transcription of the Staphylococcus aureus collagen adhesin gene (cna) in an agr-independent manner.
Mol Microbiol: 1999, 33(2);317-26
[PubMed:10411748] [WorldCat.org] [DOI] (P p)A Manna, A L Cheung
Characterization of sarR, a modulator of sar expression in Staphylococcus aureus.
Infect Immun: 2001, 69(2);885-96
[PubMed:11159982] [WorldCat.org] [DOI] (P p)B Fournier, A Klier, G Rapoport
The two-component system ArlS-ArlR is a regulator of virulence gene expression in Staphylococcus aureus.
Mol Microbiol: 2001, 41(1);247-61
[PubMed:11454217] [WorldCat.org] [DOI] (P p)M Bischoff, J M Entenza, P Giachino
Influence of a functional sigB operon on the global regulators sar and agr in Staphylococcus aureus.
J Bacteriol: 2001, 183(17);5171-9
[PubMed:11489871] [WorldCat.org] [DOI] (P p)P M Dunman, E Murphy, S Haney, D Palacios, G Tucker-Kellogg, S Wu, E L Brown, R J Zagursky, D Shlaes, S J Projan
Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci.
J Bacteriol: 2001, 183(24);7341-53
[PubMed:11717293] [WorldCat.org] [DOI] (P p)Jon S Blevins, Karen E Beenken, Mohamed O Elasri, Barry K Hurlburt, Mark S Smeltzer
Strain-dependent differences in the regulatory roles of sarA and agr in Staphylococcus aureus.
Infect Immun: 2002, 70(2);470-80
[PubMed:11796572] [WorldCat.org] [DOI] (P p)Jaione Valle, Alejandro Toledo-Arana, Carmen Berasain, Jean-Marc Ghigo, Beatriz Amorena, José R Penadés, Iñigo Lasa
SarA and not sigmaB is essential for biofilm development by Staphylococcus aureus.
Mol Microbiol: 2003, 48(4);1075-87
[PubMed:12753197] [WorldCat.org] [DOI] (P p)Jutta Rossi, Markus Bischoff, Akihito Wada, Brigitte Berger-Bächi
MsrR, a putative cell envelope-associated element involved in Staphylococcus aureus sarA attenuation.
Antimicrob Agents Chemother: 2003, 47(8);2558-64
[PubMed:12878519] [WorldCat.org] [DOI] (P p)Lindsey Shaw, Ewa Golonka, Jan Potempa, Simon J Foster
The role and regulation of the extracellular proteases of Staphylococcus aureus.
Microbiology (Reading): 2004, 150(Pt 1);217-228
[PubMed:14702415] [WorldCat.org] [DOI] (P p)Jessica O O'Leary, Mark J Langevin, Christopher T D Price, Jon S Blevins, Mark S Smeltzer, John E Gustafson
Effects of sarA inactivation on the intrinsic multidrug resistance mechanism of Staphylococcus aureus.
FEMS Microbiol Lett: 2004, 237(2);297-302
[PubMed:15321676] [WorldCat.org] [DOI] (P p)Balakrishnan Prithiviraj, Harsh P Bais, Ajay K Jha, Jorge M Vivanco
Staphylococcus aureus pathogenicity on Arabidopsis thaliana is mediated either by a direct effect of salicylic acid on the pathogen or by SA-dependent, NPR1-independent host responses.
Plant J: 2005, 42(3);417-32
[PubMed:15842626] [WorldCat.org] [DOI] (P p)María Pilar Trotonda, Adhar C Manna, Ambrose L Cheung, Iñigo Lasa, José R Penadés
SarA positively controls bap-dependent biofilm formation in Staphylococcus aureus.
J Bacteriol: 2005, 187(16);5790-8
[PubMed:16077127] [WorldCat.org] [DOI] (P p)A L Cheung, S J Projan
Cloning and sequencing of sarA of Staphylococcus aureus, a gene required for the expression of agr.
J Bacteriol: 1994, 176(13);4168-72
[PubMed:8021198] [WorldCat.org] [DOI] (P p)M G Bayer, J H Heinrichs, A L Cheung
The molecular architecture of the sar locus in Staphylococcus aureus.
J Bacteriol: 1996, 178(15);4563-70
[PubMed:8755885] [WorldCat.org] [DOI] (P p)Y Chien, A L Cheung
Molecular interactions between two global regulators, sar and agr, in Staphylococcus aureus.
J Biol Chem: 1998, 273(5);2645-52
[PubMed:9446568] [WorldCat.org] [DOI] (P p)
