⊟Summary[edit | edit source]
- organism: Staphylococcus aureus NCTC8325
- locus tag: SAOUHSC_00812
- pan locus tag?: SAUPAN002779000
- symbol: SAOUHSC_00812
- pan gene symbol?: clfA
- synonym:
- product: clumping factor
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
- type: CDS
- locus tag: SAOUHSC_00812
- symbol: SAOUHSC_00812
- product: clumping factor
- replicon: chromosome
- strand: +
- coordinates: 792986..795769
- length: 2784
- essential: no DEG other strains
⊟Accession numbers[edit | edit source]
- Gene ID: 3919375 NCBI
- RefSeq: YP_499368 NCBI
- BioCyc: G1I0R-760 BioCyc
- MicrobesOnline: 1289279 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421
481
541
601
661
721
781
841
901
961
1021
1081
1141
1201
1261
1321
1381
1441
1501
1561
1621
1681
1741
1801
1861
1921
1981
2041
2101
2161
2221
2281
2341
2401
2461
2521
2581
2641
2701
2761ATGAATATGAAGAAAAAAGAAAAACACGCAATTCGGAAAAAATCGATTGGCGTGGCTTCA
GTGCTTGTAGGTACGTTAATCGGTTTTGGACTACTCAGCAGTAAAGAAGCAGATGCAAGT
GAAAATAGTGTTACGCAATCTGATAGCGCAAGTAACGAAAGCAAAAGTAATGATTCAAGT
AGCGTTAGTGCTGCACCTAAAACAGACGACACAAACGTGAGTGATACTAAAACATCGTCA
AACACTAATAATGGCGAAACGAGTGTGGCGCAAAATCCAGCACAACAGGAAACGACACAA
TCATCATCAACAAATGCAACTACGGAAGAAACGCCGGTAACTGGTGAAGCTACTACTACG
ACAACGAATCAAGCTAATACACCGGCAACAACTCAATCAAGCAATACAAATGCGGAGGAA
TTAGTGAATCAAACAAGTAATGAAACGACTTCTAATGATACTAATACAGTATCATCTGTA
AATTCACCTCAAAATTCTACAAATGCGGAAAATGTTTCAACAACGCAAGATACTTCAACT
GAAGCAACACCTTCAAACAATGAATCAGCTCCACAGAGTACAGATGCAAGTAATAAAGAT
GTAGTTAATCAAGCGGTTAATACAAGTGCGCCTAGAATGAGAGCATTTAGTTTAGCGGCA
GTAGCTGCAGATGCACCGGTAGCTGGCACAGATATTACGAATCAGTTGACGAATGTGACA
GTTGGTATTGACTCTGGTACGACTGTGTATCCGCACCAAGCAGGTTATGTCAAACTGAAT
TATGGTTTTTCAGTGCCTAATTCTGCTGTTAAAGGTGACACATTCAAAATAACTGTACCT
AAAGAATTAAACTTAAATGGTGTAACTTCAACTGCTAAAGTGCCACCAATTATGGCTGGA
GATCAAGTATTGGCAAATGGTGTAATCGATAGTGATGGTAATGTTATTTATACATTTACA
GACTATGTAAATACTAAAGATGATGTAAAAGCAACTTTGACCATGCCCGCTTATATTGAC
CCTGAAAATGTTAAAAAGACAGGTAATGTGACATTGGCTACTGGCATAGGTAGTACAACA
GCAAACAAAACAGTATTAGTAGATTATGAAAAATATGGTAAGTTTTATAACTTATCTATT
AAAGGTACAATTGACCAAATCGATAAAACAAATAATACGTATCGTCAGACAATTTATGTC
AATCCAAGTGGAGATAACGTTATTGCGCCGGTTTTAACAGGTAATTTAAAACCAAATACG
GATAGTAATGCATTAATAGATCAGCAAAATACAAGTATTAAAGTATATAAAGTAGATAAT
GCAGCTGATTTATCTGAAAGTTACTTTGTGAATCCAGAAAACTTTGAGGATGTCACTAAT
AGTGTGAATATTACATTCCCAAATCCAAATCAATATAAAGTAGAGTTTAATACGCCTGAT
GATCAAATTACAACACCGTATATAGTAGTTGTTAATGGTCATATTGATCCGAATAGCAAA
GGTGATTTAGCTTTACGTTCAACTTTATATGGGTATAACTCGAATATAATTTGGCGCTCT
ATGTCATGGGACAACGAAGTAGCATTTAATAACGGATCAGGTTCTGGTGACGGTATCGAT
AAACCAGTTGTTCCTGAACAACCTGATGAGCCTGGTGAAATTGAACCAATTCCAGAGGAT
TCAGATTCTGACCCAGGTTCAGATTCTGGCAGCGATTCTAATTCAGATAGCGGTTCAGAT
TCGGGTAGTGATTCTACATCAGATAGTGGTTCAGATTCAGCGAGTGATTCAGATTCAGCA
AGTGATTCAGACTCAGCGAGTGATTCAGATTCAGCAAGCGATTCCGACTCAGCGAGCGAT
TCCGACTCAGACAATGACTCGGATTCAGATAGCGATTCTGACTCAGACAGTGACTCAGAT
TCCGACAGTGACTCAGATTCAGATAGCGATTCTGACTCAGACAGTGACTCGGATTCAGAT
AGCGATTCAGATTCAGATAGCGATTCAGATTCCGACAGTGATTCCGACTCAGACAGCGAT
TCTGACTCCGACAGTGATTCCGACTCAGACAGCGATTCAGATTCCGACAGTGATTCCGAC
TCAGATAGCGATTCCGACTCAGATAGCGACTCAGATTCAGACAGCGATTCAGATTCAGAC
AGCGATTCAGATTCAGATAGCGATTCAGATTCCGACAGTGACTCAGATTCCGACAGTGAC
TCGGATTCAGATAGCGATTCAGATTCCGACAGTGACTCAGATTCCGACAGTGACTCAGAC
TCAGACAGTGATTCGGATTCAGCGAGTGATTCGGATTCAGATAGTGATTCCGACTCCGAC
AGTGACTCGGATTCAGATAGCGACTCAGACTCGGATAGCGACTCGGATTCAGATAGCGAT
TCGGACTCAGATAGCGATTCAGAATCAGACAGCGATTCAGATTCAGACAGCGACTCAGAC
AGTGACTCAGATTCAGATAGTGACTCGGATTCAGCGAGTGATTCAGACTCAGGTAGTGAC
TCCGATTCATCAAGTGATTCCGACTCAGAAAGTGATTCAAATAGCGATTCCGAGTCAGTT
TCTAACAATAATGTAGTTCCGCCTAATTCACCTAAAAATGGTACTAATGCTTCTAATAAA
AATGAGGCTAAAGATAGTAAAGAACCATTACCAGATACAGGTTCTGAAGATGAAGCAAAT
ACGTCACTAATTTGGGGATTATTAGCATCAATAGGTTCATTACTACTTTTCAGAAGAAAA
AAAGAAAATAAAGATAAGAAATAA60
120
180
240
300
360
420
480
540
600
660
720
780
840
900
960
1020
1080
1140
1200
1260
1320
1380
1440
1500
1560
1620
1680
1740
1800
1860
1920
1980
2040
2100
2160
2220
2280
2340
2400
2460
2520
2580
2640
2700
2760
2784
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SAOUHSC_00812
- symbol: SAOUHSC_00812
- description: clumping factor
- length: 927
- theoretical pI: 3.44945
- theoretical MW: 96446.8
- GRAVY: -1.07206
⊟Function[edit | edit source]
- TIGRFAM: gram-positive signal peptide, YSIRK family (TIGR01168; HMM-score: 28.4)Cell envelope Other LPXTG cell wall anchor domain (TIGR01167; HMM-score: 27.7)
- TheSEED :
- Clumping factor ClfA, fibrinogen-binding protein
- PFAM: Adhesin (CL0204) SdrG_C_C; C-terminus of bacterial fibrinogen-binding adhesin (PF10425; HMM-score: 165.2)and 3 moreE-set (CL0159) Big_8; Bacterial Ig domain (PF17961; HMM-score: 90.6)no clan defined YSIRK_signal; YSIRK type signal peptide (PF04650; HMM-score: 36.6)Gram_pos_anchor; LPXTG cell wall anchor motif (PF00746; HMM-score: 23.4)
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors:
- effectors:
⊟Localization[edit | edit source]
- PSORTb: Cellwall
- Cytoplasmic Score: 0
- Cytoplasmic Membrane Score: 0
- Cellwall Score: 10
- Extracellular Score: 0
- Internal Helices: 0
- DeepLocPro: Cell wall & surface
- Cytoplasmic Score: 0
- Cytoplasmic Membrane Score: 0.0027
- Cell wall & surface Score: 0.894
- Extracellular Score: 0.1034
- LocateP: LPxTG Cell-wall anchored
- Prediction by SwissProt Classification: Cell Wall
- Pathway Prediction: Sec-(SPI)
- Intracellular possibility: 0
- Signal peptide possibility: 1
- N-terminally Anchored Score: -1
- Predicted Cleavage Site: found LPxTG motif :LPDTG
- SignalP: Signal peptide SP(Sec/SPI) length 39 aa
- SP(Sec/SPI): 0.955997
- TAT(Tat/SPI): 0.010304
- LIPO(Sec/SPII): 0.029148
- Cleavage Site: CS pos: 39-40. ADA-SE. Pr: 0.8951
- predicted transmembrane helices (TMHMM): 1
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MNMKKKEKHAIRKKSIGVASVLVGTLIGFGLLSSKEADASENSVTQSDSASNESKSNDSSSVSAAPKTDDTNVSDTKTSSNTNNGETSVAQNPAQQETTQSSSTNATTEETPVTGEATTTTTNQANTPATTQSSNTNAEELVNQTSNETTSNDTNTVSSVNSPQNSTNAENVSTTQDTSTEATPSNNESAPQSTDASNKDVVNQAVNTSAPRMRAFSLAAVAADAPVAGTDITNQLTNVTVGIDSGTTVYPHQAGYVKLNYGFSVPNSAVKGDTFKITVPKELNLNGVTSTAKVPPIMAGDQVLANGVIDSDGNVIYTFTDYVNTKDDVKATLTMPAYIDPENVKKTGNVTLATGIGSTTANKTVLVDYEKYGKFYNLSIKGTIDQIDKTNNTYRQTIYVNPSGDNVIAPVLTGNLKPNTDSNALIDQQNTSIKVYKVDNAADLSESYFVNPENFEDVTNSVNITFPNPNQYKVEFNTPDDQITTPYIVVVNGHIDPNSKGDLALRSTLYGYNSNIIWRSMSWDNEVAFNNGSGSGDGIDKPVVPEQPDEPGEIEPIPEDSDSDPGSDSGSDSNSDSGSDSGSDSTSDSGSDSASDSDSASDSDSASDSDSASDSDSASDSDSDNDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSASDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSDSESDSDSDSDSDSDSDSDSDSDSDSASDSDSGSDSDSSSDSDSESDSNSDSESVSNNNVVPPNSPKNGTNASNKNEAKDSKEPLPDTGSEDEANTSLIWGLLASIGSLLLFRRKKENKDKK
⊟Experimental data[edit | edit source]
- experimentally validated: PeptideAtlas [1] [2]
- protein localization: data available for COL
- quantitative data / protein copy number per cell:
- interaction partners:
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
- MicrobesOnline: no polycistronic organisation predicted
⊟Regulation[edit | edit source]
- regulator: SigB* (activation) regulon
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: [4]
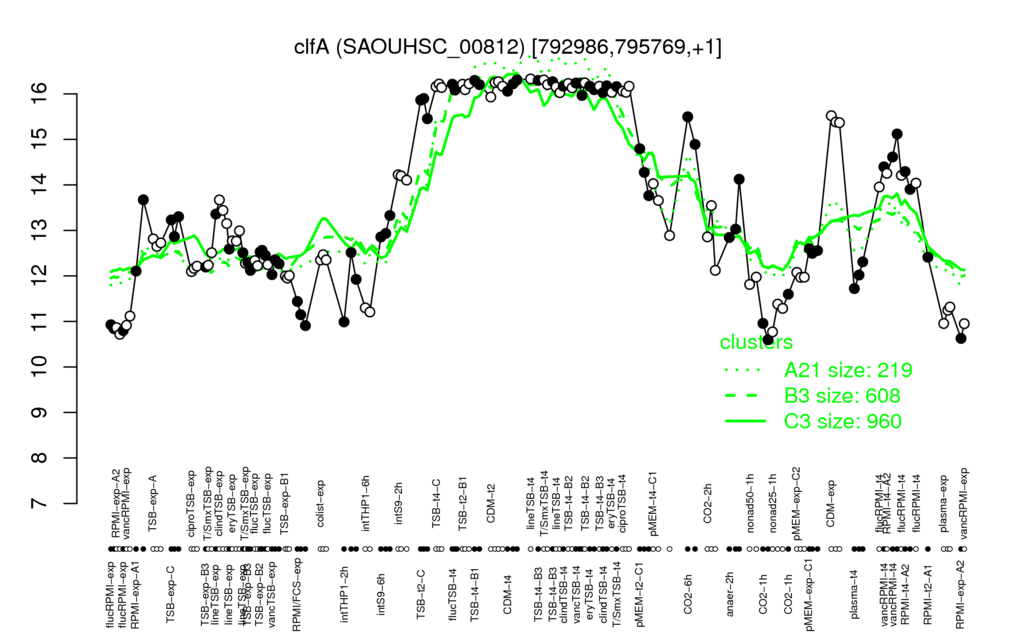 Multi-gene expression profiles
Multi-gene expression profiles
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
- Aureolib: no data available
⊟Protein stability[edit | edit source]
- half-life: no data available
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You can add further information about the gene and protein here. [edit]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ Maren Depke, Stephan Michalik, Alexander Rabe, Kristin Surmann, Lars Brinkmann, Nico Jehmlich, Jörg Bernhardt, Michael Hecker, Bernd Wollscheid, Zhi Sun, Robert L Moritz, Uwe Völker, Frank Schmidt
A peptide resource for the analysis of Staphylococcus aureus in host-pathogen interaction studies.
Proteomics: 2015, 15(21);3648-61
[PubMed:26224020] [WorldCat.org] [DOI] (I p) - ↑ Stephan Michalik, Maren Depke, Annette Murr, Manuela Gesell Salazar, Ulrike Kusebauch, Zhi Sun, Tanja C Meyer, Kristin Surmann, Henrike Pförtner, Petra Hildebrandt, Stefan Weiss, Laura Marcela Palma Medina, Melanie Gutjahr, Elke Hammer, Dörte Becher, Thomas Pribyl, Sven Hammerschmidt, Eric W Deutsch, Samuel L Bader, Michael Hecker, Robert L Moritz, Ulrike Mäder, Uwe Völker, Frank Schmidt
A global Staphylococcus aureus proteome resource applied to the in vivo characterization of host-pathogen interactions.
Sci Rep: 2017, 7(1);9718
[PubMed:28887440] [WorldCat.org] [DOI] (I e) - ↑ Markus Bischoff, Paul Dunman, Jan Kormanec, Daphne Macapagal, Ellen Murphy, William Mounts, Brigitte Berger-Bächi, Steven Projan
Microarray-based analysis of the Staphylococcus aureus sigmaB regulon.
J Bacteriol: 2004, 186(13);4085-99
[PubMed:15205410] [WorldCat.org] [DOI] (P p) - ↑ 4.0 4.1 Ulrike Mäder, Pierre Nicolas, Maren Depke, Jan Pané-Farré, Michel Debarbouille, Magdalena M van der Kooi-Pol, Cyprien Guérin, Sandra Dérozier, Aurelia Hiron, Hanne Jarmer, Aurélie Leduc, Stephan Michalik, Ewoud Reilman, Marc Schaffer, Frank Schmidt, Philippe Bessières, Philippe Noirot, Michael Hecker, Tarek Msadek, Uwe Völker, Jan Maarten van Dijl
Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions.
PLoS Genet: 2016, 12(4);e1005962
[PubMed:27035918] [WorldCat.org] [DOI] (I e)
⊟Relevant publications[edit | edit source]
T J Foster, M Höök
Surface protein adhesins of Staphylococcus aureus.
Trends Microbiol: 1998, 6(12);484-8
[PubMed:10036727] [WorldCat.org] [DOI] (P p)M E Dominiecki, J Weiss
Antibacterial action of extracellular mammalian group IIA phospholipase A2 against grossly clumped Staphylococcus aureus.
Infect Immun: 1999, 67(5);2299-305
[PubMed:10225887] [WorldCat.org] [DOI] (P p)O M Hartford, E R Wann, M Höök, T J Foster
Identification of residues in the Staphylococcus aureus fibrinogen-binding MSCRAMM clumping factor A (ClfA) that are important for ligand binding.
J Biol Chem: 2001, 276(4);2466-73
[PubMed:11044451] [WorldCat.org] [DOI] (P p)Vannakambadi K Ganesh, Jose J Rivera, Emanuel Smeds, Ya-Ping Ko, M Gabriela Bowden, Elisabeth R Wann, Shivasankarappa Gurusiddappa, J Ross Fitzgerald, Magnus Höök
A structural model of the Staphylococcus aureus ClfA-fibrinogen interaction opens new avenues for the design of anti-staphylococcal therapeutics.
PLoS Pathog: 2008, 4(11);e1000226
[PubMed:19043557] [WorldCat.org] [DOI] (I p)D McDevitt, P Francois, P Vaudaux, T J Foster
Molecular characterization of the clumping factor (fibrinogen receptor) of Staphylococcus aureus.
Mol Microbiol: 1994, 11(2);237-48
[PubMed:8170386] [WorldCat.org] [DOI] (P p)
