Jump to navigation
Jump to search
NCBI: 03-AUG-2016
⊟Summary[edit | edit source]
- organism: Staphylococcus aureus NCTC8325
- locus tag: SAOUHSC_01204
- pan locus tag?: SAUPAN003521000
- symbol: SAOUHSC_01204
- pan gene symbol?: smc
- synonym:
- product: SMC domain-containing protein
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
⊟Accession numbers[edit | edit source]
- Gene ID: 3919470 NCBI
- RefSeq: YP_499741 NCBI
- BioCyc: G1I0R-1128 BioCyc
- MicrobesOnline: 1289655 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421
481
541
601
661
721
781
841
901
961
1021
1081
1141
1201
1261
1321
1381
1441
1501
1561
1621
1681
1741
1801
1861
1921
1981
2041
2101
2161
2221
2281
2341
2401
2461
2521
2581
2641
2701
2761
2821
2881
2941
3001
3061
3121
3181
3241
3301
3361
3421
3481
3541ATGGTTTATTTAAAATCAATAGATGCCATTGGATTTAAGTCTTTTGCAGATCAAACCAAT
GTTCAATTCGATAAAGGTGTAACTGCAATTGTTGGTCCAAATGGAAGCGGTAAAAGTAAT
ATTACAGATGCTATTAAATGGGTGTTGGGTGAACAATCGGCTAAATCATTACGTGGCTCA
AAAATGGAAGATATTATCTTCTCAGGTGCAGAACATCGCAAAGCTCAAAATTATGCTGAA
GTACAGTTAAGATTAGATAATCATTCTAAAAAGCTCAGTGTTGATGAAAACGAAGTTATT
GTAACAAGAAGATTGTATCGAAGTGGTGAAAGTGAGTACTACATAAATAATGACCGTGCA
AGATTAAAAGATATTGCCGATTTATTTTTAGATTCTGGATTGGGAAAAGAAGCGTATAGC
ATTATCTCGCAAGGTAGAGTTGATGAAATACTAAATGCTAAACCAATTGATAGACGTCAA
ATTATTGAAGAATCGGCTGGTGTACTTAAATATAAAAAACGTAAGGCTGAATCATTAAAT
AAACTTGACCAAACAGAAGATAATTTAACGAGAGTAGAAGACATTTTATATGATTTGGAA
GGTCGCGTAGAACCTCTAAAAGAGGAGGCAGCTATAGCTAAAGAATATAAGACACTTTCA
CATCAAATGAAACATAGTGACATTGTAGTTACAGTGCACGATATTGATCAATATACAAAT
GACAATAGACAATTAGATCAACGTTTAAATGATTTACAAGGCCAACAAGCAAATAAAGAA
GCTGACAAGCAACGTTTAAGCCAACAAATTCAACAATATAAAGGTAAACGTCATCAACTT
GATAATGATGTTGAATCGCTTAATTATCAATTAGTAAAAGCTACGGAAGCCTTTGAAAAA
TATACGGGACAATTAAATGTTTTGGAAGAACGTAAGAAAAATCAATCTGAAACAAATGCA
CGATATGAAGAAGAACAAGAAAATTTAATGGAGCTTTTAGAAAATATATCAAATGAGATT
TCTGAAGCTCAAGATACTTATAAGTCTCTGAAAAGTAAACAAAAAGAACTCAATGCTGTC
ATTCGTGAACTTGAAGAACAACTATATGTTTCAGACGAAGCACATGATGAAAAATTGGAA
GAAATTAAAAACGAATACTATACATTAATGTCAGAGCAATCAGATGTTAACAATGATATT
CGTTTTTTAAAGCATACTATAGAAGAGAATGAGGCTAAAAAATCAAGACTAGATTCTCGA
TTAGTTGAAGTTTTTGAGCAATTGAAAGATATTCAGGGTCAAATAAAAACGACAAAAAAA
GAATATCAACAGACCAACAAAGAACTTTCTGCTGTAGATAAAGAAATTAAAAATATAGAA
AAAGACCTCACTGATACAAAAAAAGCACAAAATGAATACGAAGAGAAATTGTATCAAGCA
TATCGATATACCGAAAAAATGAAAACACGTATTGATAGTTTGGCAACGCAAGAGGAAGAA
TATACTTATTTTTTCAATGGCGTCAAACATATTTTGAAAGCTAAAAATAAAGAATTAAAG
GGTATTTATGGTGCAGTTGCGGAAATTATTGATGTGCCATCTAAATTAACTCAGGCAATT
GAAACAGCATTAGGTGCTTCATTACAACATGTCATTGTAGATTCAGAAAAAGATGGACGC
CAGGCTATTCAATTTTTAAAAGAACGTAATTTAGGTCGTGCGACGTTTTTACCATTAAAT
GTTATACAGAGTAGAGTGGTAGCGACTGATATTAAATCTATTGCTAAAGAGGCAAACGGA
TTTATTAGTATCGCTTCGGAAGCAGTTAAAGTAGCACCAGAATATCAAAATATTATCGGG
AATTTATTAGGTAATACGATTATCGTTGATCATTTAAAGCATGCAAATGAATTGGCACGT
GCGATTAAATATCGAACTCGTATTGTTACTTTGGAAGGTGATATTGTAAATCCTGGTGGT
TCTATGACTGGTGGTGGCGCTCGTAAGTCAAAAAGTATTCTGTCTCAAAAAGACGAGTTG
ACAACAATGAGACACCAATTAGAAGATTACTTGCGTCAAACAGAATCATTTGAACAACAA
TTTAAAGAGTTGAAGATAAAAAGTGATCAATTAAGTGAACTGTATTTTGAAAAAAGTCAA
AAGCATAATACACTTAAAGAGCAAGTGCATCATTTTGAAATGGAGCTCGATAGATTAACT
ACACAAGAAACACAAATAAAAAATGATCATGAAGAATTCGAATTTGAAAAAAATGATGGT
TATACGAGTGACAAAAGTCGACAAACTTTGAGTGAAAAAGAAACTTATCTAGAAAGTATT
AAAGCATCTTTAAAACGACTAGAAGATGAAATTGAACGCTACACAAAACTTTCTAAAGAA
GGTAAGGAAAGCGTTACTAAAACACAACAAACCTTACATCAGAAACAATCTGATCTTGCT
GTGGTTAAAGAGCGTATTAAAACACAACAACAGACAATAGATCGATTAAATAATCAAAAT
CAACAAACTAAACATCAATTAAAAGATGTTAAAGAAAAAATTGCATTCTTTAATTCGGAT
GAAGTGATGGGCGAACAAGCTTTTCAAAATATTAAAGATCAAATTAATGGTCAACAAGAA
ACGAGAACACGCTTATCAGATGAATTAGATAAATTGAAACAACAACGTATTGAGTTGAAT
GAACAAATCGATGCGCAAGAAGCTAAACTACAAGTTTGTCACCAAGATATTTTAGCTATC
GAAAATCACTACCAAGATATTAAAGCTGAACAATCAAAGCTAGATGTATTAATTCATCAT
GCGATAGATCATTTAAATGATGAATATCAATTGACTGTTGAACGTGCGAAATCTGAATAT
ACGAGTGATGAATCGATTGACGCATTACGTAAAAAAGTTAAGTTAATGAAGATGTCGATT
GATGAACTAGGTCCTGTAAACTTAAATGCAATTGAACAATTTGAAGAGTTAAATGAACGT
TATACATTTTTAAGTGAACAACGTACAGATCTTCGTAAAGCTAAAGAAACATTAGAGCAA
ATTATAAGTGAAATGGATCAAGAGGTTACTGAAAGATTTAAAGAAACTTTCCATGCTATT
CAAGGACATTTTACAGCTGTGTTCAAACAATTGTTTGGTGGAGGCGATGCAGAATTGCAA
TTAACTGAAGCCGATTATTTAACAGCTGGTATTGATATTGTGGTACAACCACCGGGTAAA
AAGTTGCAACATTTATCGTTACTGAGTGGTGGTGAGCGTGCATTAACTGCTATTGCTTTA
CTATTTGCAATTTTAAAAGTAAGATCTGCACCTTTTGTTATATTAGATGAGGTTGAAGCT
GCACTAGATGAAGCAAATGTTATTAGATACGCAAAATATTTAAATGAGTTATCAGACGAA
ACACAATTCATTGTTATTACACACCGTAAAGGAACAATGGAATTTGCAGATAGGTTATAC
GGTGTAACAATGCAAGAATCAGGTGTTACTAAACTTGTGAGTGTGAATTTAAATACAATA
GATGATGTGTTGAAGGAGGAGCAATAA60
120
180
240
300
360
420
480
540
600
660
720
780
840
900
960
1020
1080
1140
1200
1260
1320
1380
1440
1500
1560
1620
1680
1740
1800
1860
1920
1980
2040
2100
2160
2220
2280
2340
2400
2460
2520
2580
2640
2700
2760
2820
2880
2940
3000
3060
3120
3180
3240
3300
3360
3420
3480
3540
3567
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SAOUHSC_01204
- symbol: SAOUHSC_01204
- description: SMC domain-containing protein
- length: 1188
- theoretical pI: 5.14611
- theoretical MW: 136748
- GRAVY: -0.759428
⊟Function[edit | edit source]
- TIGRFAM: Cellular processes Cell division chromosome segregation protein SMC (TIGR02168; HMM-score: 1104)DNA metabolism Chromosome-associated proteins chromosome segregation protein SMC (TIGR02168; HMM-score: 1104)and 24 moreCellular processes Cell division chromosome segregation protein SMC (TIGR02169; HMM-score: 659.4)DNA metabolism Chromosome-associated proteins chromosome segregation protein SMC (TIGR02169; HMM-score: 659.4)DNA metabolism DNA replication, recombination, and repair DNA repair protein RecN (TIGR00634; HMM-score: 55.8)helix-rich protein (TIGR04523; HMM-score: 44.7)thiol reductant ABC exporter, CydD subunit (TIGR02857; HMM-score: 34.9)Cell envelope Biosynthesis and degradation of surface polysaccharides and lipopolysaccharides lipid A export permease/ATP-binding protein MsbA (TIGR02203; HMM-score: 32)Transport and binding proteins Other lipid A export permease/ATP-binding protein MsbA (TIGR02203; HMM-score: 32)ABC transporter, permease/ATP-binding protein (TIGR02204; HMM-score: 31.2)DNA metabolism DNA replication, recombination, and repair exonuclease SbcC (TIGR00618; HMM-score: 30.3)Transport and binding proteins Cations and iron carrying compounds cobalt ABC transporter, ATP-binding protein (TIGR01166; HMM-score: 29.8)Cellular processes Pathogenesis type I secretion system ATPase (TIGR03375; HMM-score: 27.6)Protein fate Protein and peptide secretion and trafficking type I secretion system ATPase (TIGR03375; HMM-score: 27.6)Transport and binding proteins Carbohydrates, organic alcohols, and acids glucan exporter ATP-binding protein (TIGR01192; EC 3.6.3.42; HMM-score: 27.3)Transport and binding proteins Amino acids, peptides and amines urea ABC transporter, ATP-binding protein UrtD (TIGR03411; HMM-score: 26.9)Transport and binding proteins Cations and iron carrying compounds nickel import ATP-binding protein NikD (TIGR02770; HMM-score: 24.3)SH3 domain protein (TIGR04211; HMM-score: 21.9)proposed F420-0 ABC transporter, ATP-binding protein (TIGR03873; HMM-score: 18.4)Transport and binding proteins Other antigen peptide transporter 2 (TIGR00958; HMM-score: 17.4)Protein fate Protein and peptide secretion and trafficking type I secretion system ATPase (TIGR01842; HMM-score: 14)thiol reductant ABC exporter, CydC subunit (TIGR02868; HMM-score: 13.1)Protein fate Protein and peptide secretion and trafficking heme ABC exporter, ATP-binding protein CcmA (TIGR01189; EC 3.6.3.41; HMM-score: 12.6)Transport and binding proteins Other heme ABC exporter, ATP-binding protein CcmA (TIGR01189; EC 3.6.3.41; HMM-score: 12.6)DNA metabolism Restriction/modification DNA sulfur modification protein DndD (TIGR03185; HMM-score: 10.9)Hypothetical proteins Conserved TIGR02680 family protein (TIGR02680; HMM-score: 7.3)
- TheSEED :
- Chromosome partition protein smc
- PFAM: P-loop_NTPase (CL0023) SMC_N; RecF/RecN/SMC N terminal domain (PF02463; HMM-score: 212.3)and 11 moreno clan defined SMC_hinge; SMC proteins Flexible Hinge Domain (PF06470; HMM-score: 111.6)P-loop_NTPase (CL0023) AAA_23; AAA domain (PF13476; HMM-score: 78.6)AAA_15; AAA ATPase domain (PF13175; HMM-score: 44.8)AAA_21; AAA domain, putative AbiEii toxin, Type IV TA system (PF13304; HMM-score: 31.8)AAA_29; P-loop containing region of AAA domain (PF13555; HMM-score: 29.8)SbcC_Walker_B; SbcC/RAD50-like, Walker B motif (PF13558; HMM-score: 13.5)no clan defined DUF6026; Family of unknown function (DUF6026) (PF19491; HMM-score: 9.7)Fez1; Fez1 (PF06818; HMM-score: 9.3)DUF730; Protein of unknown function (DUF730) (PF05325; HMM-score: 8.2)DUF1664; Protein of unknown function (DUF1664) (PF07889; HMM-score: 8.1)P-loop_NTPase (CL0023) AAA_22; AAA domain (PF13401; HMM-score: 8)
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors:
- effectors:
⊟Localization[edit | edit source]
- PSORTb: Cytoplasmic
- Cytoplasmic Score: 9.97
- Cytoplasmic Membrane Score: 0
- Cellwall Score: 0.01
- Extracellular Score: 0.02
- Internal Helices: 0
- DeepLocPro: Cytoplasmic
- Cytoplasmic Score: 0.9891
- Cytoplasmic Membrane Score: 0.0075
- Cell wall & surface Score: 0.0022
- Extracellular Score: 0.0011
- LocateP: Intracellular
- Prediction by SwissProt Classification: Cytoplasmic
- Pathway Prediction: No pathway
- Intracellular possibility: 1
- Signal peptide possibility: -1
- N-terminally Anchored Score: -1
- Predicted Cleavage Site: No CleavageSite
- SignalP: no predicted signal peptide
- SP(Sec/SPI): 0.012867
- TAT(Tat/SPI): 0.00074
- LIPO(Sec/SPII): 0.001805
- predicted transmembrane helices (TMHMM): 0
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MVYLKSIDAIGFKSFADQTNVQFDKGVTAIVGPNGSGKSNITDAIKWVLGEQSAKSLRGSKMEDIIFSGAEHRKAQNYAEVQLRLDNHSKKLSVDENEVIVTRRLYRSGESEYYINNDRARLKDIADLFLDSGLGKEAYSIISQGRVDEILNAKPIDRRQIIEESAGVLKYKKRKAESLNKLDQTEDNLTRVEDILYDLEGRVEPLKEEAAIAKEYKTLSHQMKHSDIVVTVHDIDQYTNDNRQLDQRLNDLQGQQANKEADKQRLSQQIQQYKGKRHQLDNDVESLNYQLVKATEAFEKYTGQLNVLEERKKNQSETNARYEEEQENLMELLENISNEISEAQDTYKSLKSKQKELNAVIRELEEQLYVSDEAHDEKLEEIKNEYYTLMSEQSDVNNDIRFLKHTIEENEAKKSRLDSRLVEVFEQLKDIQGQIKTTKKEYQQTNKELSAVDKEIKNIEKDLTDTKKAQNEYEEKLYQAYRYTEKMKTRIDSLATQEEEYTYFFNGVKHILKAKNKELKGIYGAVAEIIDVPSKLTQAIETALGASLQHVIVDSEKDGRQAIQFLKERNLGRATFLPLNVIQSRVVATDIKSIAKEANGFISIASEAVKVAPEYQNIIGNLLGNTIIVDHLKHANELARAIKYRTRIVTLEGDIVNPGGSMTGGGARKSKSILSQKDELTTMRHQLEDYLRQTESFEQQFKELKIKSDQLSELYFEKSQKHNTLKEQVHHFEMELDRLTTQETQIKNDHEEFEFEKNDGYTSDKSRQTLSEKETYLESIKASLKRLEDEIERYTKLSKEGKESVTKTQQTLHQKQSDLAVVKERIKTQQQTIDRLNNQNQQTKHQLKDVKEKIAFFNSDEVMGEQAFQNIKDQINGQQETRTRLSDELDKLKQQRIELNEQIDAQEAKLQVCHQDILAIENHYQDIKAEQSKLDVLIHHAIDHLNDEYQLTVERAKSEYTSDESIDALRKKVKLMKMSIDELGPVNLNAIEQFEELNERYTFLSEQRTDLRKAKETLEQIISEMDQEVTERFKETFHAIQGHFTAVFKQLFGGGDAELQLTEADYLTAGIDIVVQPPGKKLQHLSLLSGGERALTAIALLFAILKVRSAPFVILDEVEAALDEANVIRYAKYLNELSDETQFIVITHRKGTMEFADRLYGVTMQESGVTKLVSVNLNTIDDVLKEEQ
⊟Experimental data[edit | edit source]
- experimentally validated: PeptideAtlas [2] [3]
- protein localization: data available for COL
- quantitative data / protein copy number per cell:
- interaction partners:
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
- MicrobesOnline: acpP > rnc > SAOUHSC_01204 > SAOUHSC_01205 > SAOUHSC_01206 > SAOUHSC_01207predicted SigA promoter [4] : SAOUHSC_01196 > SAOUHSC_01197 > SAOUHSC_01198 > SAOUHSC_01199 > S494 > S495 > acpP > S496 > rnc > S497 > SAOUHSC_01204 > SAOUHSC_01205 > SAOUHSC_01206 > SAOUHSC_01207 > S498predicted SigA promoter [4] : S495 > acpP > S496 > rnc > S497 > SAOUHSC_01204 > SAOUHSC_01205 > SAOUHSC_01206 > SAOUHSC_01207 > S498
⊟Regulation[edit | edit source]
- regulator:
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: [4]
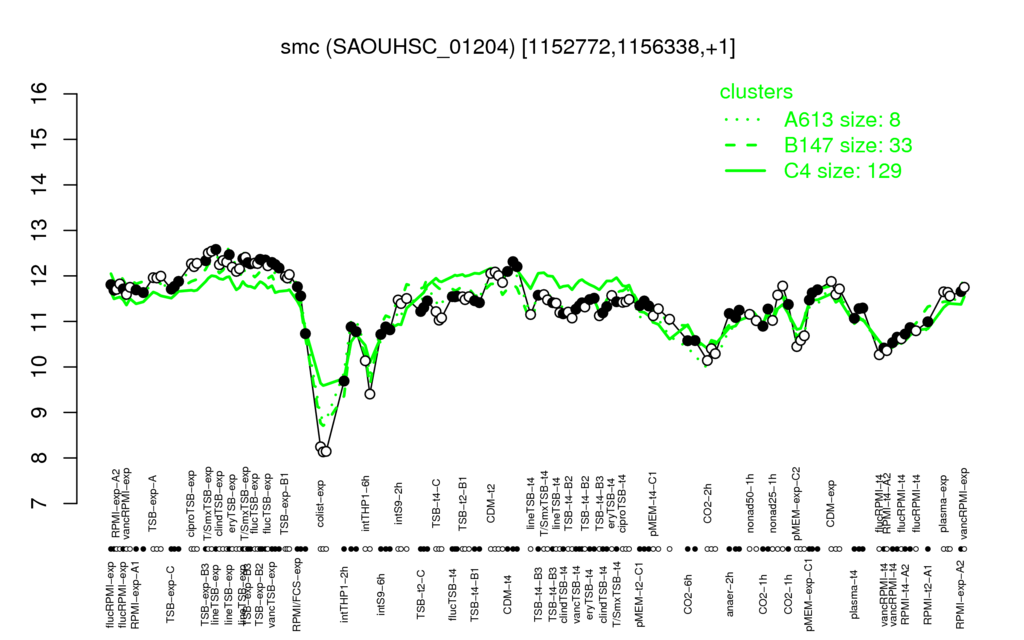 Multi-gene expression profiles
Multi-gene expression profiles
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
- Aureolib: no data available
⊟Protein stability[edit | edit source]
- half-life: no data available
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You can add further information about the gene and protein here. [edit]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ Roy R Chaudhuri, Andrew G Allen, Paul J Owen, Gil Shalom, Karl Stone, Marcus Harrison, Timothy A Burgis, Michael Lockyer, Jorge Garcia-Lara, Simon J Foster, Stephen J Pleasance, Sarah E Peters, Duncan J Maskell, Ian G Charles
Comprehensive identification of essential Staphylococcus aureus genes using Transposon-Mediated Differential Hybridisation (TMDH).
BMC Genomics: 2009, 10;291
[PubMed:19570206] [WorldCat.org] [DOI] (I e) - ↑ Maren Depke, Stephan Michalik, Alexander Rabe, Kristin Surmann, Lars Brinkmann, Nico Jehmlich, Jörg Bernhardt, Michael Hecker, Bernd Wollscheid, Zhi Sun, Robert L Moritz, Uwe Völker, Frank Schmidt
A peptide resource for the analysis of Staphylococcus aureus in host-pathogen interaction studies.
Proteomics: 2015, 15(21);3648-61
[PubMed:26224020] [WorldCat.org] [DOI] (I p) - ↑ Stephan Michalik, Maren Depke, Annette Murr, Manuela Gesell Salazar, Ulrike Kusebauch, Zhi Sun, Tanja C Meyer, Kristin Surmann, Henrike Pförtner, Petra Hildebrandt, Stefan Weiss, Laura Marcela Palma Medina, Melanie Gutjahr, Elke Hammer, Dörte Becher, Thomas Pribyl, Sven Hammerschmidt, Eric W Deutsch, Samuel L Bader, Michael Hecker, Robert L Moritz, Ulrike Mäder, Uwe Völker, Frank Schmidt
A global Staphylococcus aureus proteome resource applied to the in vivo characterization of host-pathogen interactions.
Sci Rep: 2017, 7(1);9718
[PubMed:28887440] [WorldCat.org] [DOI] (I e) - ↑ 4.0 4.1 4.2 Ulrike Mäder, Pierre Nicolas, Maren Depke, Jan Pané-Farré, Michel Debarbouille, Magdalena M van der Kooi-Pol, Cyprien Guérin, Sandra Dérozier, Aurelia Hiron, Hanne Jarmer, Aurélie Leduc, Stephan Michalik, Ewoud Reilman, Marc Schaffer, Frank Schmidt, Philippe Bessières, Philippe Noirot, Michael Hecker, Tarek Msadek, Uwe Völker, Jan Maarten van Dijl
Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions.
PLoS Genet: 2016, 12(4);e1005962
[PubMed:27035918] [WorldCat.org] [DOI] (I e)
