⊟Summary[edit | edit source]
- organism: Staphylococcus aureus NCTC8325
- locus tag: SAOUHSC_01419
- pan locus tag?: SAUPAN003833000
- symbol: SAOUHSC_01419
- pan gene symbol?: arlS
- synonym:
- product: hypothetical protein
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
- type: CDS
- locus tag: SAOUHSC_01419
- symbol: SAOUHSC_01419
- product: hypothetical protein
- replicon: chromosome
- strand: -
- coordinates: 1360281..1361636
- length: 1356
- essential: no DEG other strains
⊟Accession numbers[edit | edit source]
- Gene ID: 3920650 NCBI
- RefSeq: YP_499945 NCBI
- BioCyc: G1I0R-1324 BioCyc
- MicrobesOnline: 1289859 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421
481
541
601
661
721
781
841
901
961
1021
1081
1141
1201
1261
1321ATGACAAAACGTAAATTGCGCAATAACTGGATTATTGTTACCACGATGATTACGTTTGTC
ACGATATTTTTGTTTTGTTTAATTATTATTTTTTTCTTGAAAGATACACTGCATAATAGT
GAGCTTGATGATGCAGAACGAAGCTCAAGCGATATTAATAATTTATTTCATTCTAAGCCT
GTTAAAGATATATCTGCATTAGACTTGAATGCATCTTTAGGTAATTTTCAAGAGATAATT
ATTTATGATGAGCATAATAATAAATTATTTGAGACATCGAATGATAACACAGTGAGAGTT
GAACCAGGTTATGAACACCGTTATTTTGACCGCGTAATAAAAAAACGCTATAAAGGCATT
GAATATTTAATTATTAAAGAACCAATTACAACGCAAGATTTCAAAGGGTATAGCTTGTTA
ATTCATTCACTAGAAAATTATGATAACATCGTAAAATCATTGTATATCATTGCGCTGGCA
TTTGGAGTGATTGCAACAATTATAACTGCCACAATCAGTTATGTATTTTCAACACAAATT
ACTAAACCGCTTGTCAGTTTATCAAATAAAATGATTGAGATTCGACGAGATGGTTTTCAA
AATAAATTGCAATTAAATACAAATTATGAAGAAATAGATAATTTAGCAAATACGTTTAAT
GAGATGATGAGCCAAATTGAAGAATCATTTAATCAACAAAGACAATTTGTTGAAGATGCG
TCACATGAATTACGAACACCATTACAAATTATTCAAGGTCATTTAAATTTGATTCAGCGA
TGGGGAAAAAAAGACCCAGCAGTATTAGAAGAATCGTTAAATATTTCTATTGAAGAAATG
AATCGTATCATAAAATTAGTCGAAGAATTACTTGAATTGACTAAAGGAGATGTAAATGAC
ATTTCTTCTGAAGCACAGACCGTGCATATTAATGATGAAATTCGCTCGCGAATACACTCA
TTAAAACAATTGCATCCTGATTATCAATTTGATACGGATCTGACATCTAAAAATCTAGAA
ATTAAAATGAAACCTCATCAATTCGAACAATTATTTTTAATCTTTATTGATAATGCAATC
AAATATGATGTGAAGAATAAGAAAATTAAAGTTAAGACAAGGTTAAAAAATAAGCAAAAA
ATAATTGAAATTACAGATCATGGAATTGGTATTCCAGAGGAAGATCAAGATTTCATTTTT
GATCGCTTTTATCGAGTGGATAAATCTCGTTCAAGAAGTCAAGGCGGTAATGGACTCGGA
TTATCTATTGCTCAAAAAATCATTCAATTAAACGGAGGATCGATTAAAATTAAAAGTGAA
ATTAACAAAGGAACAACGTTTAAAATCATATTTTAA60
120
180
240
300
360
420
480
540
600
660
720
780
840
900
960
1020
1080
1140
1200
1260
1320
1356
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SAOUHSC_01419
- symbol: SAOUHSC_01419
- description: hypothetical protein
- length: 451
- theoretical pI: 6.97701
- theoretical MW: 52399.7
- GRAVY: -0.33592
⊟Function[edit | edit source]
- reaction: EC 2.7.13.3? ExPASyHistidine kinase ATP + protein L-histidine = ADP + protein N-phospho-L-histidine
- TIGRFAM: heavy metal sensor kinase (TIGR01386; EC 2.7.13.3; HMM-score: 197.2)Signal transduction Two-component systems phosphate regulon sensor kinase PhoR (TIGR02966; EC 2.7.3.-; HMM-score: 177.3)and 8 moreproteobacterial dedicated sortase system histidine kinase (TIGR03785; HMM-score: 91)Signal transduction Two-component systems TMAO reductase sytem sensor TorS (TIGR02956; EC 2.7.13.3; HMM-score: 89)Protein fate Protein and peptide secretion and trafficking putative PEP-CTERM system histidine kinase (TIGR02916; EC 2.7.13.3; HMM-score: 51.5)Signal transduction Two-component systems putative PEP-CTERM system histidine kinase (TIGR02916; EC 2.7.13.3; HMM-score: 51.5)Central intermediary metabolism Nitrogen fixation nitrogen fixation negative regulator NifL (TIGR02938; HMM-score: 49.9)Regulatory functions Protein interactions nitrogen fixation negative regulator NifL (TIGR02938; HMM-score: 49.9)Cellular processes Sporulation and germination anti-sigma F factor (TIGR01925; EC 2.7.11.1; HMM-score: 15.1)Regulatory functions Protein interactions anti-sigma F factor (TIGR01925; EC 2.7.11.1; HMM-score: 15.1)
- TheSEED :
- Two-component system histidine kinase ArlS
- PFAM: His_Kinase_A (CL0025) HATPase_c; Histidine kinase-, DNA gyrase B-, and HSP90-like ATPase (PF02518; HMM-score: 93.2)and 16 moreHisKA; His Kinase A (phospho-acceptor) domain (PF00512; HMM-score: 60)Cache (CL0165) ArlS_N; ArlS sensor domain (PF18719; HMM-score: 37.1)His_Kinase_A (CL0025) HATPase_c_3; Histidine kinase-, DNA gyrase B-, and HSP90-like ATPase (PF13589; HMM-score: 23.6)HATPase_c_5; GHKL domain (PF14501; HMM-score: 22.8)HAMP (CL0681) HAMP; HAMP domain (PF00672; HMM-score: 18.9)TPR (CL0020) COG6_N; Conserved oligomeric complex COG6, N-terminal (PF06419; HMM-score: 18.9)no clan defined DUF2207_C; Predicted membrane protein (DUF2207) C-terminal domain (PF20990; HMM-score: 15.9)TSPAN_4TM-like (CL0347) Tetraspanin; Tetraspanin family (PF00335; HMM-score: 15)no clan defined Carbpep_Y_N; Carboxypeptidase Y pro-peptide (PF05388; HMM-score: 14.2)ZapG-like; Z-ring associated protein G-like (PF06295; HMM-score: 13.9)MukF_C; MukF C-terminal domain (PF17193; HMM-score: 12.8)Clusterin; Clusterin (PF01093; HMM-score: 12)DUF1664; Protein of unknown function (DUF1664) (PF07889; HMM-score: 9.9)DUF2232; Predicted membrane protein (DUF2232) (PF09991; HMM-score: 9)DUF4133; Domain of unknown function (DUF4133) (PF13571; HMM-score: 8.9)DUF948; Bacterial protein of unknown function (DUF948) (PF06103; HMM-score: 7.3)
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors:
- effectors:
⊟Localization[edit | edit source]
- PSORTb: Cytoplasmic Membrane
- Cytoplasmic Score: 0.17
- Cytoplasmic Membrane Score: 9.51
- Cellwall Score: 0.16
- Extracellular Score: 0.15
- Internal Helices: 2
- DeepLocPro: Cytoplasmic Membrane
- Cytoplasmic Score: 0
- Cytoplasmic Membrane Score: 0.9986
- Cell wall & surface Score: 0.0001
- Extracellular Score: 0.0012
- LocateP: Multi-transmembrane
- Prediction by SwissProt Classification: Membrane
- Pathway Prediction: Sec-(SPI)
- Intracellular possibility: 0.17
- Signal peptide possibility: -0.5
- N-terminally Anchored Score: 1
- Predicted Cleavage Site: No CleavageSite
- SignalP: no predicted signal peptide
- SP(Sec/SPI): 0.009038
- TAT(Tat/SPI): 0.000301
- LIPO(Sec/SPII): 0.004974
- predicted transmembrane helices (TMHMM): 2
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MTKRKLRNNWIIVTTMITFVTIFLFCLIIIFFLKDTLHNSELDDAERSSSDINNLFHSKPVKDISALDLNASLGNFQEIIIYDEHNNKLFETSNDNTVRVEPGYEHRYFDRVIKKRYKGIEYLIIKEPITTQDFKGYSLLIHSLENYDNIVKSLYIIALAFGVIATIITATISYVFSTQITKPLVSLSNKMIEIRRDGFQNKLQLNTNYEEIDNLANTFNEMMSQIEESFNQQRQFVEDASHELRTPLQIIQGHLNLIQRWGKKDPAVLEESLNISIEEMNRIIKLVEELLELTKGDVNDISSEAQTVHINDEIRSRIHSLKQLHPDYQFDTDLTSKNLEIKMKPHQFEQLFLIFIDNAIKYDVKNKKIKVKTRLKNKQKIIEITDHGIGIPEEDQDFIFDRFYRVDKSRSRSQGGNGLGLSIAQKIIQLNGGSIKIKSEINKGTTFKIIF
⊟Experimental data[edit | edit source]
- experimentally validated: PeptideAtlas [1] [2]
- protein localization: data available for COL
- quantitative data / protein copy number per cell:
- interaction partners:
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
⊟Regulation[edit | edit source]
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: [3]
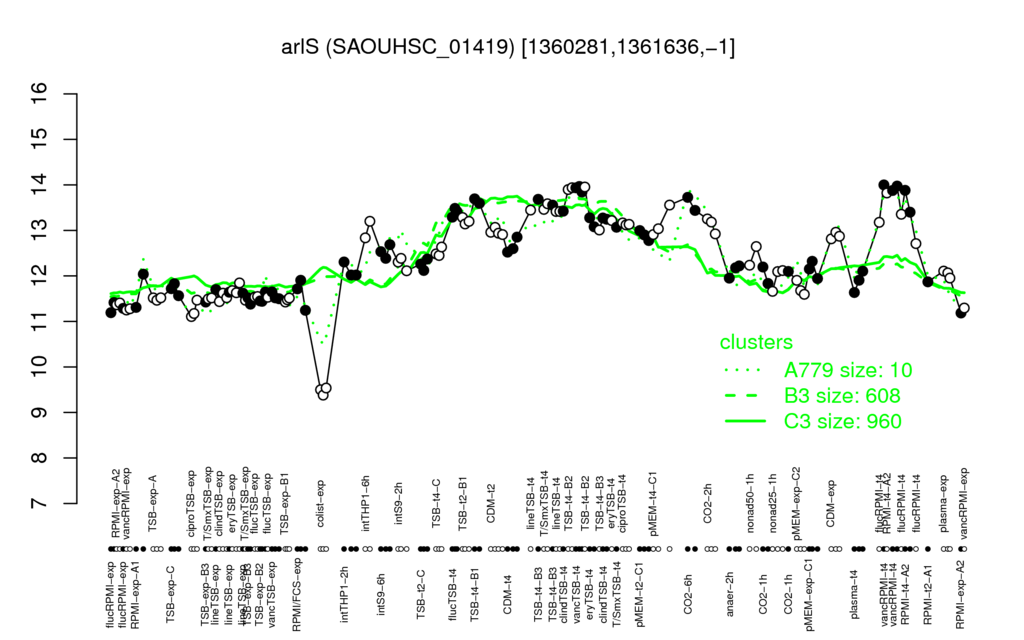 Multi-gene expression profiles
Multi-gene expression profiles
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
- Aureolib: no data available
⊟Protein stability[edit | edit source]
- half-life: no data available
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You can add further information about the gene and protein here. [edit]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ Maren Depke, Stephan Michalik, Alexander Rabe, Kristin Surmann, Lars Brinkmann, Nico Jehmlich, Jörg Bernhardt, Michael Hecker, Bernd Wollscheid, Zhi Sun, Robert L Moritz, Uwe Völker, Frank Schmidt
A peptide resource for the analysis of Staphylococcus aureus in host-pathogen interaction studies.
Proteomics: 2015, 15(21);3648-61
[PubMed:26224020] [WorldCat.org] [DOI] (I p) - ↑ Stephan Michalik, Maren Depke, Annette Murr, Manuela Gesell Salazar, Ulrike Kusebauch, Zhi Sun, Tanja C Meyer, Kristin Surmann, Henrike Pförtner, Petra Hildebrandt, Stefan Weiss, Laura Marcela Palma Medina, Melanie Gutjahr, Elke Hammer, Dörte Becher, Thomas Pribyl, Sven Hammerschmidt, Eric W Deutsch, Samuel L Bader, Michael Hecker, Robert L Moritz, Ulrike Mäder, Uwe Völker, Frank Schmidt
A global Staphylococcus aureus proteome resource applied to the in vivo characterization of host-pathogen interactions.
Sci Rep: 2017, 7(1);9718
[PubMed:28887440] [WorldCat.org] [DOI] (I e) - ↑ 3.0 3.1 Ulrike Mäder, Pierre Nicolas, Maren Depke, Jan Pané-Farré, Michel Debarbouille, Magdalena M van der Kooi-Pol, Cyprien Guérin, Sandra Dérozier, Aurelia Hiron, Hanne Jarmer, Aurélie Leduc, Stephan Michalik, Ewoud Reilman, Marc Schaffer, Frank Schmidt, Philippe Bessières, Philippe Noirot, Michael Hecker, Tarek Msadek, Uwe Völker, Jan Maarten van Dijl
Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions.
PLoS Genet: 2016, 12(4);e1005962
[PubMed:27035918] [WorldCat.org] [DOI] (I e)
⊟Relevant publications[edit | edit source]
B Fournier, R Aras, D C Hooper
Expression of the multidrug resistance transporter NorA from Staphylococcus aureus is modified by a two-component regulatory system.
J Bacteriol: 2000, 182(3);664-71
[PubMed:10633099] [WorldCat.org] [DOI] (P p)B Fournier, D C Hooper
A new two-component regulatory system involved in adhesion, autolysis, and extracellular proteolytic activity of Staphylococcus aureus.
J Bacteriol: 2000, 182(14);3955-64
[PubMed:10869073] [WorldCat.org] [DOI] (P p)B Fournier, A Klier, G Rapoport
The two-component system ArlS-ArlR is a regulator of virulence gene expression in Staphylococcus aureus.
Mol Microbiol: 2001, 41(1);247-61
[PubMed:11454217] [WorldCat.org] [DOI] (P p)S S Ingavale, W Van Wamel, A L Cheung
Characterization of RAT, an autolysis regulator in Staphylococcus aureus.
Mol Microbiol: 2003, 48(6);1451-66
[PubMed:12791130] [WorldCat.org] [DOI] (P p)Adhar C Manna, Susham S Ingavale, MaryBeth Maloney, Willem van Wamel, Ambrose L Cheung
Identification of sarV (SA2062), a new transcriptional regulator, is repressed by SarA and MgrA (SA0641) and involved in the regulation of autolysis in Staphylococcus aureus.
J Bacteriol: 2004, 186(16);5267-80
[PubMed:15292128] [WorldCat.org] [DOI] (P p)Bénédicte Fournier, André Klier
Protein A gene expression is regulated by DNA supercoiling which is modified by the ArlS-ArlR two-component system of Staphylococcus aureus.
Microbiology (Reading): 2004, 150(Pt 11);3807-3819
[PubMed:15528666] [WorldCat.org] [DOI] (P p)Alejandro Toledo-Arana, Nekane Merino, Marta Vergara-Irigaray, Michel Débarbouillé, José R Penadés, Iñigo Lasa
Staphylococcus aureus develops an alternative, ica-independent biofilm in the absence of the arlRS two-component system.
J Bacteriol: 2005, 187(15);5318-29
[PubMed:16030226] [WorldCat.org] [DOI] (P p)
