Jump to navigation
Jump to search
NCBI: 03-AUG-2016
⊟Summary[edit | edit source]
- organism: Staphylococcus aureus NCTC8325
- locus tag: SAOUHSC_02401
- pan locus tag?: SAUPAN005496000
- symbol: SAOUHSC_02401
- pan gene symbol?: mtlR
- synonym:
- product: hypothetical protein
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
- type: CDS
- locus tag: SAOUHSC_02401
- symbol: SAOUHSC_02401
- product: hypothetical protein
- replicon: chromosome
- strand: +
- coordinates: 2221120..2223252
- length: 2133
- essential: no DEG other strains
- comment: The sequence of SAOUHSC_02401 was corrected based on the resequencing performed by Berscheid et al., 2012 [1].
⊟Accession numbers[edit | edit source]
- Gene ID: 3919616 NCBI
- RefSeq: YP_500877 NCBI
- BioCyc: G1I0R-2267 BioCyc
- MicrobesOnline: 1290838 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421
481
541
601
661
721
781
841
901
961
1021
1081
1141
1201
1261
1321
1381
1441
1501
1561
1621
1681
1741
1801
1861
1921
1981
2041
2101ATGTTATTGAGTACACGTGAAAAAGAAATGATAGCCCTATTGATTAAGTACCACGGTCAA
TATATCACTATACACGACATTGCTCAGCAACTTGCGGTGTCCTCTCGTACTATTCACCGT
GAATTAAAAGGTGTTGAAGCATATTTAACTTCATTTTCATTAACTTTAGAACGCGCAAAC
AAAAAAGGGCTACGCATTGCTGGCACAGATTCTGATTTAAACGATTTGAAGCAATCGATT
GCACAACATCAAACCATTGACTTATCTGTTGAAGAGCAGAAAGTAATTATTATATACGCT
TTGATACAAGCCAAGGAGCCAGTTAAACAATATAGTTTAGCGCAAGAAATTGGCGTTTCT
GTCCAGACTTTAGCAAAGATGTTAGATGATTTAGAGCTTGATTTAAATAAGTACCAACTA
TCTTTATCTCGAAAGCGTGGCGAAGGCATTTACTTGGTAGGTACTGAATCAAAGAAACGT
GAATTTTTAAGTCAATTAATGGTGAATAACTTAAATAGTACTAGCGTTTATTCAGTAATT
GAAAATCATTTTGTCTTTCATTCATTAAATCAAATCCACAAAGACTTTGTTGACTTAGAG
CGCATTTTTAATGTTGAAAGACTATTAATGGACTACCTAAGTGCCTTACCCTACCAACTT
ACCGAATCAAGTTATTTAACTTTAACTGTCCATATCGTGCTCTCCATTTCACGTATAAAA
AATGGAGAGTATGTCGCATTAAACGATGATATTTATGATTCTGTACAAAACACATTTGAA
CACAAAGTAGCAAGCGAACTTGCTGATAAACTTGGTCAAATATATGACGTCACGTTTAAT
CAGGCAGAAATTGCTTTCATTACTATCCATTTACGTGGAGCTAAACGAAAAAATCTTAAT
GATACATCATTAAATAATCGTTGTGAAGAAAACAAAATTAAAGCGTTTGTTAACAAAGTA
GAAATGATTTCCGGTATGACATTTGCAGATTTGGATACTTTAGTAGATGGACTGACGCTA
CACCTTAATCCTGCAATCAATCGTTTGCAAGCTAATATCGAGACCTATAATCCGTTAACA
GACATGATTAAGTTCAAATATCCAAGACTATTTGAAAATGTAAGATTAGCTTTAAATGAT
TGTTGGCCTGATTTGATTTTTCCAGAGAGTGAAATTGCTTTTATAGTTTTACACTTTGGT
GGCTCGATTAAAAACCAAGGTAATCGATTTTTAAACATATTAGTCGTTTGCAGCAGTGGT
ATGGGAACTAGTCGTCTATTATCAACTCGTCTAGAGCAAGTTTTTAGTGAGATTGAGCGT
ATTACACAAGCATCAGTCAGCGATTTGAAGTCACTAGATTTAAGTCAATATGATGGCATT
ATTTCTACTGTGAATTTAGACATCGACTCCCCCTATTTAACGGTAAACCCATTATTACCA
GATAGTGATATCAGTTATGTCGCACAGTTTTTAAATACAAAGTCTACGTTCCAAGAGACG
CATGATAAATCATCAAACATGATTGATAAGGATGATGTTCATGTTGAAACGAAAGATGTT
GATGGCAACACATCTTTTGAAAATGAACAAACTTCATACTTAACTTCAGTTTTCGAAAAA
CATTTAAGTGACGAAAAATCAGAACAATTATTGCATCATATGCGTTCGGGTTTAACTTTG
CTTGATTCAGTGAAAATAGTTAGTACCGAAGTTAAACAGTGGCAAACATATATCGCAGAT
TATCTATATCAATGCGATGTAATAAACGATCCAACGTCATTCGCTGAACTACTAGAGCAA
CGATTGATTGACAATCCAGGATGGATATTAAGTCCATATCCTGTTGCAATACCACACCTA
AGAGACAATATGATTAAACACCCTATGATTCTAATCACAGTTTTAGAAGAACCGTTAACA
TTGCCTAGTATTCAAAATGACAATCAAACAATTAAATATATGATTTCCATGTTTATTTCT
GACAATGATTTTATGGCATCACTGGTAAGTGACTTGTCCGAATTTTTAAGTTTGAAATTA
GAATCTATTGATACTTTTATGGAAAATCCACAGGAACTTGAAACATTATTAAGAAACAAA
TTTTTAGAACGAATTAAAAAACAATTTATTTAG60
120
180
240
300
360
420
480
540
600
660
720
780
840
900
960
1020
1080
1140
1200
1260
1320
1380
1440
1500
1560
1620
1680
1740
1800
1860
1920
1980
2040
2100
2133
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SAOUHSC_02401
- symbol: SAOUHSC_02401
- description: hypothetical protein
- length: 710
- theoretical pI: 4.88526
- theoretical MW: 81145.1
- GRAVY: -0.175493
⊟Function[edit | edit source]
- TIGRFAM: Transport and binding proteins Carbohydrates, organic alcohols, and acids PTS system, fructose subfamily, IIA component (TIGR00848; EC 2.7.1.69; HMM-score: 28)Signal transduction PTS PTS system, fructose subfamily, IIA component (TIGR00848; EC 2.7.1.69; HMM-score: 28)and 9 moreBiosynthesis of cofactors, prosthetic groups, and carriers Other iron-sulfur cluster biosynthesis transcriptional regulator SufR (TIGR02702; HMM-score: 16.3)Regulatory functions DNA interactions iron-sulfur cluster biosynthesis transcriptional regulator SufR (TIGR02702; HMM-score: 16.3)RNA polymerase sigma-70 factor, sigma-B/F/G subfamily (TIGR02980; HMM-score: 16)RNA polymerase sigma factor, TIGR02999 family (TIGR02999; HMM-score: 13.6)RNA polymerase sigma factor, sigma-70 family (TIGR02937; HMM-score: 13)Unknown function Enzymes of unknown specificity NDMA-dependent alcohol dehydrogenase, Rxyl_3153 family (TIGR03989; EC 1.1.99.36; HMM-score: 12.9)Transport and binding proteins Carbohydrates, organic alcohols, and acids PTS system, lactose/cellobiose family IIB component (TIGR00853; HMM-score: 12.7)Signal transduction PTS PTS system, lactose/cellobiose family IIB component (TIGR00853; HMM-score: 12.7)probable regulatory domain (TIGR03879; HMM-score: 12.6)
- TheSEED :
- Mannitol operon activator, BglG family
- PFAM: PRD (CL0166) PRD; PRD domain (PF00874; HMM-score: 122.7)and 23 moreHTH (CL0123) Mga; Mga helix-turn-helix domain (PF05043; HMM-score: 55.7)HTH_11; HTH domain (PF08279; HMM-score: 49.3)PTase-anion_tr (CL0340) PTS_EIIA_2; Phosphoenolpyruvate-dependent sugar phosphotransferase system, EIIA 2 (PF00359; HMM-score: 36.1)Phosphatase (CL0031) PTS_IIB; PTS system, Lactose/Cellobiose specific IIB subunit (PF02302; HMM-score: 29.5)HTH (CL0123) HTH_24; Winged helix-turn-helix DNA-binding (PF13412; HMM-score: 19.5)Fe_dep_repress; Iron dependent repressor, N-terminal DNA binding domain (PF01325; HMM-score: 18.4)HTH_23; Homeodomain-like domain (PF13384; HMM-score: 17.2)HTH_38; Helix-turn-helix domain (PF13936; HMM-score: 17.1)HTH_10; HTH DNA binding domain (PF04967; HMM-score: 16.9)MarR; MarR family (PF01047; HMM-score: 16.7)IF2_N; Translation initiation factor IF-2, N-terminal region (PF04760; HMM-score: 16.7)HTH_DeoR; DeoR-like helix-turn-helix domain (PF08220; HMM-score: 15.5)HTH_Crp_2; Crp-like helix-turn-helix domain (PF13545; HMM-score: 15.4)MarR_2; MarR family (PF12802; HMM-score: 15)GntR; Bacterial regulatory proteins, gntR family (PF00392; HMM-score: 13.4)HTH_Mga; M protein trans-acting positive regulator (MGA) HTH domain (PF08280; HMM-score: 13)no clan defined Phage_T7_55; Bacteriophage T7, gene 5.5 (PF11247; HMM-score: 13)HTH (CL0123) HTH_7; Helix-turn-helix domain of resolvase (PF02796; HMM-score: 12.5)HTH_Tnp_ISL3; Helix-turn-helix domain of transposase family ISL3 (PF13542; HMM-score: 12.5)Sigma70_r4; Sigma-70, region 4 (PF04545; HMM-score: 11.9)RuvB_C; RuvB C-terminal winged helix domain (PF05491; HMM-score: 11)TPR (CL0020) UVSSA_N; UVSSA N-terminal domain (PF20867; HMM-score: 11)HTH (CL0123) HTH_Tnp_1_2; Helix-turn-helix of insertion element transposase (PF13022; HMM-score: 7.7)
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors:
- effectors: HPr (phosphocarrier protein); MtlA (mannitol-specific enzyme IICB PTS component)
- genes regulated by MtlR*, TF important in Mannitol utilizationRegPrecisetranscription unit transferred from N315 data RegPrecise
⊟Localization[edit | edit source]
- PSORTb: Cytoplasmic
- Cytoplasmic Score: 7.5
- Cytoplasmic Membrane Score: 1.15
- Cellwall Score: 0.62
- Extracellular Score: 0.73
- Internal Helices: 0
- DeepLocPro: Cytoplasmic
- Cytoplasmic Score: 0.8926
- Cytoplasmic Membrane Score: 0.1026
- Cell wall & surface Score: 0.0004
- Extracellular Score: 0.0043
- LocateP: Intracellular
- Prediction by SwissProt Classification: Cytoplasmic
- Pathway Prediction: No pathway
- Intracellular possibility: 1
- Signal peptide possibility: -1
- N-terminally Anchored Score: 1
- Predicted Cleavage Site: No CleavageSite
- SignalP: no predicted signal peptide
- SP(Sec/SPI): 0.003666
- TAT(Tat/SPI): 0.00113
- LIPO(Sec/SPII): 0.000625
- predicted transmembrane helices (TMHMM): 0
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MLLSTREKEMIALLIKYHGQYITIHDIAQQLAVSSRTIHRELKGVEAYLTSFSLTLERANKKGLRIAGTDSDLNDLKQSIAQHQTIDLSVEEQKVIIIYALIQAKEPVKQYSLAQEIGVSVQTLAKMLDDLELDLNKYQLSLSRKRGEGIYLVGTESKKREFLSQLMVNNLNSTSVYSVIENHFVFHSLNQIHKDFVDLERIFNVERLLMDYLSALPYQLTESSYLTLTVHIVLSISRIKNGEYVALNDDIYDSVQNTFEHKVASELADKLGQIYDVTFNQAEIAFITIHLRGAKRKNLNDTSLNNRCEENKIKAFVNKVEMISGMTFADLDTLVDGLTLHLNPAINRLQANIETYNPLTDMIKFKYPRLFENVRLALNDCWPDLIFPESEIAFIVLHFGGSIKNQGNRFLNILVVCSSGMGTSRLLSTRLEQVFSEIERITQASVSDLKSLDLSQYDGIISTVNLDIDSPYLTVNPLLPDSDISYVAQFLNTKSTFQETHDKSSNMIDKDDVHVETKDVDGNTSFENEQTSYLTSVFEKHLSDEKSEQLLHHMRSGLTLLDSVKIVSTEVKQWQTYIADYLYQCDVINDPTSFAELLEQRLIDNPGWILSPYPVAIPHLRDNMIKHPMILITVLEEPLTLPSIQNDNQTIKYMISMFISDNDFMASLVSDLSEFLSLKLESIDTFMENPQELETLLRNKFLERIKKQFI
⊟Experimental data[edit | edit source]
- experimentally validated: PeptideAtlas [2] [3]
- protein localization: data available for COL
- quantitative data / protein copy number per cell:
- interaction partners:
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
⊟Regulation[edit | edit source]
- regulators: SigB* (activation) regulon, MtlR* (activation) regulon
SigB* (sigma factor) controlling a large regulon involved in stress/starvation response and adaptation; [5] [4] other strains MtlR* (TF) important in Mannitol utilization; RegPrecise transcription unit transferred from N315 data RegPrecise
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: [4]
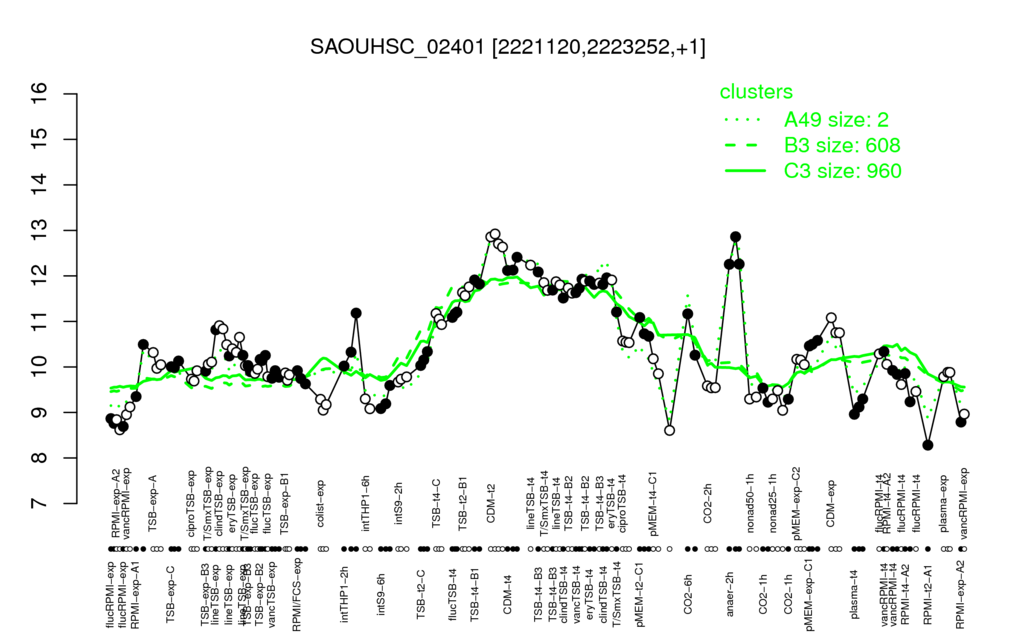 Multi-gene expression profiles
Multi-gene expression profiles
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
- Aureolib: no data available
⊟Protein stability[edit | edit source]
- half-life: no data available
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You can add further information about the gene and protein here. [edit]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ Anne Berscheid, Peter Sass, Konstantin Weber-Lassalle, Ambrose L Cheung, Gabriele Bierbaum
Revisiting the genomes of the Staphylococcus aureus strains NCTC 8325 and RN4220.
Int J Med Microbiol: 2012, 302(2);84-7
[PubMed:22417616] [WorldCat.org] [DOI] (I p) - ↑ Maren Depke, Stephan Michalik, Alexander Rabe, Kristin Surmann, Lars Brinkmann, Nico Jehmlich, Jörg Bernhardt, Michael Hecker, Bernd Wollscheid, Zhi Sun, Robert L Moritz, Uwe Völker, Frank Schmidt
A peptide resource for the analysis of Staphylococcus aureus in host-pathogen interaction studies.
Proteomics: 2015, 15(21);3648-61
[PubMed:26224020] [WorldCat.org] [DOI] (I p) - ↑ Stephan Michalik, Maren Depke, Annette Murr, Manuela Gesell Salazar, Ulrike Kusebauch, Zhi Sun, Tanja C Meyer, Kristin Surmann, Henrike Pförtner, Petra Hildebrandt, Stefan Weiss, Laura Marcela Palma Medina, Melanie Gutjahr, Elke Hammer, Dörte Becher, Thomas Pribyl, Sven Hammerschmidt, Eric W Deutsch, Samuel L Bader, Michael Hecker, Robert L Moritz, Ulrike Mäder, Uwe Völker, Frank Schmidt
A global Staphylococcus aureus proteome resource applied to the in vivo characterization of host-pathogen interactions.
Sci Rep: 2017, 7(1);9718
[PubMed:28887440] [WorldCat.org] [DOI] (I e) - ↑ 4.0 4.1 4.2 Ulrike Mäder, Pierre Nicolas, Maren Depke, Jan Pané-Farré, Michel Debarbouille, Magdalena M van der Kooi-Pol, Cyprien Guérin, Sandra Dérozier, Aurelia Hiron, Hanne Jarmer, Aurélie Leduc, Stephan Michalik, Ewoud Reilman, Marc Schaffer, Frank Schmidt, Philippe Bessières, Philippe Noirot, Michael Hecker, Tarek Msadek, Uwe Völker, Jan Maarten van Dijl
Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions.
PLoS Genet: 2016, 12(4);e1005962
[PubMed:27035918] [WorldCat.org] [DOI] (I e) - ↑ Markus Bischoff, Paul Dunman, Jan Kormanec, Daphne Macapagal, Ellen Murphy, William Mounts, Brigitte Berger-Bächi, Steven Projan
Microarray-based analysis of the Staphylococcus aureus sigmaB regulon.
J Bacteriol: 2004, 186(13);4085-99
[PubMed:15205410] [WorldCat.org] [DOI] (P p)
