Jump to navigation
Jump to search
NCBI: 03-AUG-2016
⊟Summary[edit | edit source]
- organism: Staphylococcus aureus NCTC8325
- locus tag: SAOUHSC_02873
- pan locus tag?: SAUPAN006220000
- symbol: SAOUHSC_02873
- pan gene symbol?: copA
- synonym:
- product: cation transporter E1-E2 family ATPase
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
- type: CDS
- locus tag: SAOUHSC_02873
- symbol: SAOUHSC_02873
- product: cation transporter E1-E2 family ATPase
- replicon: chromosome
- strand: +
- coordinates: 2645181..2647589
- length: 2409
- essential: no DEG other strains
⊟Accession numbers[edit | edit source]
- Gene ID: 3921544 NCBI
- RefSeq: YP_501328 NCBI
- BioCyc: G1I0R-2705 BioCyc
- MicrobesOnline: 1291299 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421
481
541
601
661
721
781
841
901
961
1021
1081
1141
1201
1261
1321
1381
1441
1501
1561
1621
1681
1741
1801
1861
1921
1981
2041
2101
2161
2221
2281
2341
2401GTGGCTAATACGAAAAAAACAACATTAGATATCACTGGTATGACTTGTGCCGCATGTTCA
AATCGTATCGAAAAGAAACTGAATAAACTTGATGACGTTAATGCCCAAGTGAATTTAACT
ACAGAGAAAGCAACTGTTGAGTATAACCCTGATCAACATGATGTCCAAGAATTTATTAAT
ACGATTCAACATTTAGGTTACGGTGTCGCTGTAGAAACTGTCGAATTAGACATTACAGGT
ATGACTTGTGCTGCATGCTCAAGCCGTATTGAAAAAGTGTTAAATAAAATGGACGGCGTT
CAAAATGCAACGGTCAATTTAACAACAGAGCAAGCTAAAGTTGACTATTATCCTGAAGAA
ACAGATGCTGATAAACTTGTCACTCGCATTCAAAAATTAGGTTATGACGCGTCTATTAAA
GATAACAATAAAGATCAAACGTCACGCAAAGCTGAAGCGCTACAACATAAATTGATTAAG
CTTATCATATCAGCAGTATTATCTTTACCACTATTAATGTTAATGTTTGTACATCTTTTC
AATATGCATATACCAGCACTATTTACGAATCCATGGTTCCAATTTATTTTAGCTACACCT
GTACAATTTATTATTGGATGGCAATTTTATGTAGGTGCTTATAAAAACTTAAGAAATGGT
GGCGCCAATATGGATGTACTTGTTGCTGTTGGTACAAGTGCAGCATATTTTTACAGTATT
TATGAAATGGTTCGTTGGCTAAATGGCTCAACAACGCAACCGCATTTATACTTTGAAACA
AGCGCCGTACTAATTACCTTAATCTTATTCGGTAAGTATTTAGAAGCTAGAGCGAAGTCT
CAAACAACCAATGCGCTTGGCGAATTATTAAGTTTACAAGCTAAAGAAGCACGCATTTTA
AAAGATGGTAATGAAGTGATGATTCCTCTAAATGAAGTACATGTTGGAGATACACTTATC
GTTAAACCAGGTGAAAAGATACCTGTTGATGGCAAAATTATTAAAGGTATGACTGCCATC
GACGAATCTATGTTAACAGGTGAATCTATCCCTGTTGAGAAGAATGTTGATGATACTGTA
ATTGGTTCAACGATGAACAAAAACGGTACTATTACTATGACAGCAACAAAAGTTGGCGGG
GACACTGCGTTGGCAAATATTATTAAAGTTGTCGAAGAAGCTCAAAGTTCTAAAGCGCCG
ATTCAACGATTGGCAGATATTATTTCTGGTTATTTCGTTCCTATCGTTGTTGGTATCGCA
CTATTAACATTTATCGTGTGGATTACTTTAGTTACACCAGGTACATTTGAACCTGCACTT
GTTGCGAGTATTTCCGTTCTCGTCATTGCTTGTCCATGCGCATTGGGACTTGCTACACCA
ACTTCTATTATGGTAGGTACTGGTCGCGCTGCTGAAAATGGTATTTTATTTAAAGGTGGC
GAGTTTGTTGAACGCACACATCAAATTGATACCATCGTTTTAGATAAGACGGGTACCATT
ACAAATGGTCGTCCAGTCGTGACAGATTATCATGGTGACAATCAAACGCTACAACTACTT
GCTACTGCTGAAAAAGATTCTGAACACCCATTGGCAGAAGCCATTGTCAATTATGCAAAA
GAAAAGCAATTAATATTAACTGAGACAACAACATTTAAAGCAGTACCTGGCCATGGTATT
GAAGCAACGATTGATCATCACCATATATTGGTTGGTAACCGTAAATTAATGGCTGACAAT
GATATTAGCTTGCCTAAGCATATTTCTGATGATTTAACACATTATGAACGAGATGGTAAA
ACTGCTATGCTCATTGCTGTTAATTATTCATTAACTGGTATCATCGCAGTGGCAGATACT
GTCAAAGATCATGCCAAAGATGCTATAAAACAATTGCATGATATGGGCATTGAAGTTGCC
ATGTTAACTGGCGATAATAAAAACACTGCTCAAGCCATTGCAAAACAAGTAGGCATAGAT
ACTGTTATTGCAGATATTTTACCAGAAGAAAAAGCTGCACAAATTGCGAAACTACAGCAA
CAAGGTAAGAAGGTTGCGATGGTTGGTGACGGTGTAAATGATGCACCTGCATTAGTTAAA
GCTGATATCGGTATCGCCATTGGTACAGGTACAGAAGTTGCCATTGAAGCAGCTGATATT
ACTATTCTTGGTGGCGACTTGATGCTTATTCCTAAAGCCATTTATGCAAGTAAAGCAACC
ATTCGTAATATTCGTCAAAATCTATTTTGGGCATTCGGCTATAATATTGCCGGTATCCCT
ATAGCTGCATTGGGCTTACTTGCGCCATGGGTTGCTGGTGCTGCAATGGCACTAAGTTCA
GTAAGTGTTGTCACAAACGCACTTAGATTGAAAAAGATGCGATTAGAACCACGCCGTAAA
GATGCCTAG60
120
180
240
300
360
420
480
540
600
660
720
780
840
900
960
1020
1080
1140
1200
1260
1320
1380
1440
1500
1560
1620
1680
1740
1800
1860
1920
1980
2040
2100
2160
2220
2280
2340
2400
2409
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SAOUHSC_02873
- symbol: SAOUHSC_02873
- description: cation transporter E1-E2 family ATPase
- length: 802
- theoretical pI: 6.15296
- theoretical MW: 86743.8
- GRAVY: 0.107481
⊟Function[edit | edit source]
- reaction: EC 3.6.3.54? ExPASyCu+ exporting ATPase ATP + H2O + Cu+(Side 1) = ADP + phosphate + Cu+(Side 2)
- TIGRFAM: heavy metal translocating P-type ATPase (TIGR01525; EC 3.6.3.-; HMM-score: 671.6)Cellular processes Detoxification copper-translocating P-type ATPase (TIGR01511; EC 3.6.3.4; HMM-score: 633.6)Transport and binding proteins Cations and iron carrying compounds copper-translocating P-type ATPase (TIGR01511; EC 3.6.3.4; HMM-score: 633.6)and 31 moreTransport and binding proteins Cations and iron carrying compounds cadmium-translocating P-type ATPase (TIGR01512; EC 3.6.3.3; HMM-score: 426.6)HAD ATPase, P-type, family IC (TIGR01494; EC 3.6.3.-; HMM-score: 326.1)Transport and binding proteins Cations and iron carrying compounds K+-transporting ATPase, B subunit (TIGR01497; EC 3.6.3.12; HMM-score: 203.5)plasma-membrane proton-efflux P-type ATPase (TIGR01647; EC 3.6.3.6; HMM-score: 186)calcium-translocating P-type ATPase, PMCA-type (TIGR01517; EC 3.6.3.8; HMM-score: 159.7)calcium-transporting P-type ATPase, PMR1-type (TIGR01522; EC 3.6.3.8; HMM-score: 136.1)Energy metabolism ATP-proton motive force interconversion Na,H/K antiporter P-type ATPase, alpha subunit (TIGR01106; EC 3.6.3.-; HMM-score: 133.9)Transport and binding proteins Cations and iron carrying compounds calcium-translocating P-type ATPase, SERCA-type (TIGR01116; EC 3.6.3.8; HMM-score: 132.2)Transport and binding proteins Cations and iron carrying compounds magnesium-translocating P-type ATPase (TIGR01524; EC 3.6.3.2; HMM-score: 105.8)P-type ATPase of unknown pump specificity (type V) (TIGR01657; HMM-score: 102)potassium/sodium efflux P-type ATPase, fungal-type (TIGR01523; EC 3.6.3.-; HMM-score: 100)Transport and binding proteins Cations and iron carrying compounds copper ion binding protein (TIGR00003; HMM-score: 74.3)Cellular processes Detoxification mercuric transport protein periplasmic component (TIGR02052; HMM-score: 63.8)Unknown function Enzymes of unknown specificity Cof-like hydrolase (TIGR00099; HMM-score: 38.3)Amino acid biosynthesis Serine family phosphoserine phosphatase SerB (TIGR00338; EC 3.1.3.3; HMM-score: 29.3)phospholipid-translocating P-type ATPase, flippase (TIGR01652; EC 3.6.3.1; HMM-score: 28.1)phosphoserine phosphatase-like hydrolase, archaeal (TIGR01491; HMM-score: 24.1)Cell envelope Biosynthesis and degradation of surface polysaccharides and lipopolysaccharides 3-deoxy-D-manno-octulosonate 8-phosphate phosphatase, YrbI family (TIGR01670; EC 3.1.3.45; HMM-score: 23.6)phosphoglycolate phosphatase, TA0175-type (TIGR01487; EC 3.1.3.18; HMM-score: 22.9)Unknown function Enzymes of unknown specificity HAD phosphoserine phosphatase-like hydrolase, family IB (TIGR01488; HMM-score: 21.3)sucrose-phosphate phosphatase subfamily (TIGR01482; EC 3.1.3.-; HMM-score: 19.2)mannosyl-3-phosphoglycerate phosphatase family (TIGR01486; EC 3.1.3.-; HMM-score: 19.1)phosphonatase-like hydrolase (TIGR03351; HMM-score: 18.4)HAD hydrolase, TIGR01548 family (TIGR01548; HMM-score: 14.7)Unknown function Enzymes of unknown specificity HAD hydrolase, family IB (TIGR01490; HMM-score: 13.1)Cellular processes Pathogenesis type III secretion apparatus lipoprotein, YscJ/HrcJ family (TIGR02544; HMM-score: 11.6)Protein fate Protein and peptide secretion and trafficking type III secretion apparatus lipoprotein, YscJ/HrcJ family (TIGR02544; HMM-score: 11.6)Biosynthesis of cofactors, prosthetic groups, and carriers Other Fe-S cluster assembly protein NifU (TIGR02000; HMM-score: 11.2)Central intermediary metabolism Nitrogen fixation Fe-S cluster assembly protein NifU (TIGR02000; HMM-score: 11.2)Cellular processes Cell division chromosome segregation protein SMC (TIGR02169; HMM-score: 9.9)DNA metabolism Chromosome-associated proteins chromosome segregation protein SMC (TIGR02169; HMM-score: 9.9)
- TheSEED :
- Copper-translocating P-type ATPase (EC 3.6.3.4)
- Lead, cadmium, zinc and mercury transporting ATPase (EC 3.6.3.3) (EC 3.6.3.5)
Respiration Electron accepting reactions Terminal cytochrome C oxidases Copper-translocating P-type ATPase (EC 3.6.3.4)and 1 more - PFAM: no clan defined E1-E2_ATPase; E1-E2 ATPase (PF00122; HMM-score: 199)and 19 moreHAD (CL0137) Hydrolase; haloacid dehalogenase-like hydrolase (PF00702; HMM-score: 138.2)HMA (CL0704) HMA; Heavy-metal-associated domain (PF00403; HMM-score: 123.4)HAD (CL0137) Hydrolase_3; haloacid dehalogenase-like hydrolase (PF08282; HMM-score: 29)HAD; haloacid dehalogenase-like hydrolase (PF12710; HMM-score: 23.5)TusA-like (CL0397) TusA; Sulfurtransferase TusA (PF01206; HMM-score: 20.1)HAD (CL0137) Hydrolase_6; haloacid dehalogenase-like hydrolase (PF13344; HMM-score: 19.4)HAD_2; haloacid dehalogenase-like hydrolase (PF13419; HMM-score: 16.9)no clan defined DUF6286; Family of unknown function (DUF6286) (PF19803; HMM-score: 14.7)DUF7573; Domain of unknown function (DUF7573) (PF24458; HMM-score: 14.4)DNM3A_N; DNA (cytosine-5-)-methyltransferase, N-terminal (PF22855; HMM-score: 14.1)PIN (CL0280) PIN_9; PIN like domain (PF18477; HMM-score: 13.9)Glyoxalase (CL0104) YycE-like_C; YycE-like C-terminal domain (PF22659; HMM-score: 13.8)RNase_H (CL0219) TNP-like_RNaseH_N; TNP-like, RNase H N-terminal domain (PF21787; HMM-score: 13.5)GlnB-like (CL0089) DUF190; Uncharacterized ACR, COG1993 (PF02641; HMM-score: 13.1)no clan defined DUF2730; Protein of unknown function (DUF2730) (PF10805; HMM-score: 13.1)HMA (CL0704) HMA_2; Heavy metal associated domain 2 (PF19991; HMM-score: 13)Zn_Beta_Ribbon (CL0167) Zn_ribbon_DUF2089; DUF2089 zinc ribbon (PF22747; HMM-score: 11.9)no clan defined Phage_holin_3_6; Putative Actinobacterial Holin-X, holin superfamily III (PF07332; HMM-score: 9.8)Spore_YhcN_YlaJ; Sporulation lipoprotein YhcN/YlaJ (Spore_YhcN_YlaJ) (PF09580; HMM-score: 8.6)
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors:
- effectors:
⊟Localization[edit | edit source]
- PSORTb: Cytoplasmic Membrane
- Cytoplasmic Score: 0
- Cytoplasmic Membrane Score: 9.99
- Cellwall Score: 0.01
- Extracellular Score: 0.01
- Internal Helices: 8
- DeepLocPro: Cytoplasmic Membrane
- Cytoplasmic Score: 0
- Cytoplasmic Membrane Score: 0.9998
- Cell wall & surface Score: 0
- Extracellular Score: 0.0002
- LocateP: Multi-transmembrane
- Prediction by SwissProt Classification: Membrane
- Pathway Prediction: Sec-(SPI)
- Intracellular possibility: 0.17
- Signal peptide possibility: -1
- N-terminally Anchored Score: 1
- Predicted Cleavage Site: No CleavageSite
- SignalP: no predicted signal peptide
- SP(Sec/SPI): 0.017136
- TAT(Tat/SPI): 0.000966
- LIPO(Sec/SPII): 0.390678
- predicted transmembrane helices (TMHMM): 8
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MANTKKTTLDITGMTCAACSNRIEKKLNKLDDVNAQVNLTTEKATVEYNPDQHDVQEFINTIQHLGYGVAVETVELDITGMTCAACSSRIEKVLNKMDGVQNATVNLTTEQAKVDYYPEETDADKLVTRIQKLGYDASIKDNNKDQTSRKAEALQHKLIKLIISAVLSLPLLMLMFVHLFNMHIPALFTNPWFQFILATPVQFIIGWQFYVGAYKNLRNGGANMDVLVAVGTSAAYFYSIYEMVRWLNGSTTQPHLYFETSAVLITLILFGKYLEARAKSQTTNALGELLSLQAKEARILKDGNEVMIPLNEVHVGDTLIVKPGEKIPVDGKIIKGMTAIDESMLTGESIPVEKNVDDTVIGSTMNKNGTITMTATKVGGDTALANIIKVVEEAQSSKAPIQRLADIISGYFVPIVVGIALLTFIVWITLVTPGTFEPALVASISVLVIACPCALGLATPTSIMVGTGRAAENGILFKGGEFVERTHQIDTIVLDKTGTITNGRPVVTDYHGDNQTLQLLATAEKDSEHPLAEAIVNYAKEKQLILTETTTFKAVPGHGIEATIDHHHILVGNRKLMADNDISLPKHISDDLTHYERDGKTAMLIAVNYSLTGIIAVADTVKDHAKDAIKQLHDMGIEVAMLTGDNKNTAQAIAKQVGIDTVIADILPEEKAAQIAKLQQQGKKVAMVGDGVNDAPALVKADIGIAIGTGTEVAIEAADITILGGDLMLIPKAIYASKATIRNIRQNLFWAFGYNIAGIPIAALGLLAPWVAGAAMALSSVSVVTNALRLKKMRLEPRRKDA
⊟Experimental data[edit | edit source]
- experimentally validated: PeptideAtlas [1] [2]
- protein localization: data available for COL
- quantitative data / protein copy number per cell:
- interaction partners:
SAOUHSC_00799 (eno) phosphopyruvate hydratase [3] (data from MRSA252) SAOUHSC_02494 (rpsE) 30S ribosomal protein S5 [3] (data from MRSA252) SAOUHSC_00679 hypothetical protein [3] (data from MRSA252)
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
⊟Regulation[edit | edit source]
- regulator:
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: [4]
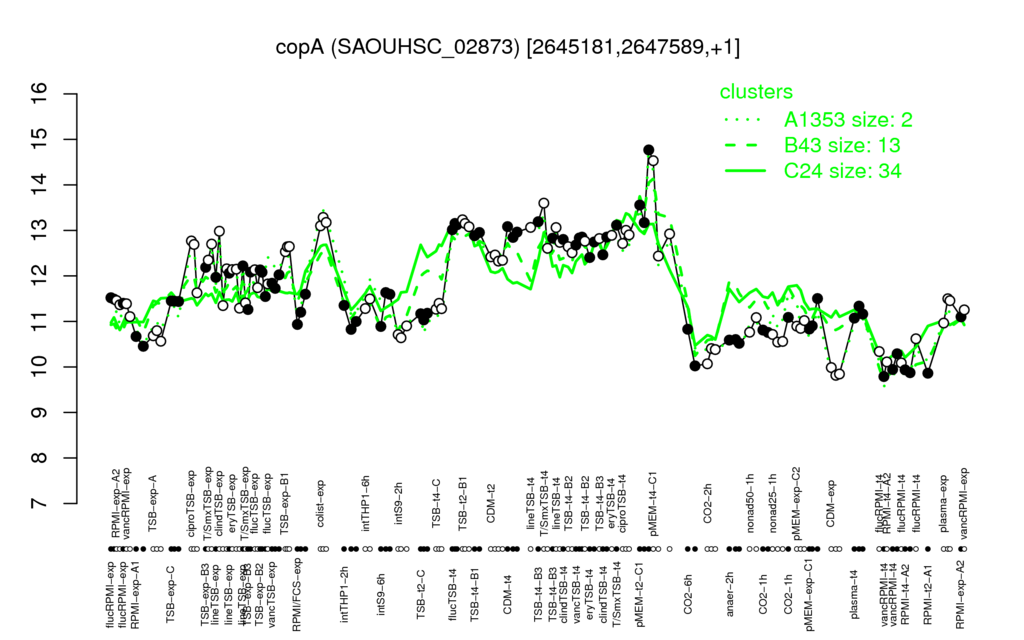 Multi-gene expression profiles
Multi-gene expression profiles
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
- Aureolib: no data available
⊟Protein stability[edit | edit source]
- half-life: no data available
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You can add further information about the gene and protein here. [edit]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ Maren Depke, Stephan Michalik, Alexander Rabe, Kristin Surmann, Lars Brinkmann, Nico Jehmlich, Jörg Bernhardt, Michael Hecker, Bernd Wollscheid, Zhi Sun, Robert L Moritz, Uwe Völker, Frank Schmidt
A peptide resource for the analysis of Staphylococcus aureus in host-pathogen interaction studies.
Proteomics: 2015, 15(21);3648-61
[PubMed:26224020] [WorldCat.org] [DOI] (I p) - ↑ Stephan Michalik, Maren Depke, Annette Murr, Manuela Gesell Salazar, Ulrike Kusebauch, Zhi Sun, Tanja C Meyer, Kristin Surmann, Henrike Pförtner, Petra Hildebrandt, Stefan Weiss, Laura Marcela Palma Medina, Melanie Gutjahr, Elke Hammer, Dörte Becher, Thomas Pribyl, Sven Hammerschmidt, Eric W Deutsch, Samuel L Bader, Michael Hecker, Robert L Moritz, Ulrike Mäder, Uwe Völker, Frank Schmidt
A global Staphylococcus aureus proteome resource applied to the in vivo characterization of host-pathogen interactions.
Sci Rep: 2017, 7(1);9718
[PubMed:28887440] [WorldCat.org] [DOI] (I e) - ↑ 3.0 3.1 3.2 Artem Cherkasov, Michael Hsing, Roya Zoraghi, Leonard J Foster, Raymond H See, Nikolay Stoynov, Jihong Jiang, Sukhbir Kaur, Tian Lian, Linda Jackson, Huansheng Gong, Rick Swayze, Emily Amandoron, Farhad Hormozdiari, Phuong Dao, Cenk Sahinalp, Osvaldo Santos-Filho, Peter Axerio-Cilies, Kendall Byler, William R McMaster, Robert C Brunham, B Brett Finlay, Neil E Reiner
Mapping the protein interaction network in methicillin-resistant Staphylococcus aureus.
J Proteome Res: 2011, 10(3);1139-50
[PubMed:21166474] [WorldCat.org] [DOI] (I p) - ↑ 4.0 4.1 Ulrike Mäder, Pierre Nicolas, Maren Depke, Jan Pané-Farré, Michel Debarbouille, Magdalena M van der Kooi-Pol, Cyprien Guérin, Sandra Dérozier, Aurelia Hiron, Hanne Jarmer, Aurélie Leduc, Stephan Michalik, Ewoud Reilman, Marc Schaffer, Frank Schmidt, Philippe Bessières, Philippe Noirot, Michael Hecker, Tarek Msadek, Uwe Völker, Jan Maarten van Dijl
Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions.
PLoS Genet: 2016, 12(4);e1005962
[PubMed:27035918] [WorldCat.org] [DOI] (I e)
⊟Relevant publications[edit | edit source]
Sutthirat Sitthisak, Lawrence Knutsson, James W Webb, Radheshyam K Jayaswal
Molecular characterization of the copper transport system in Staphylococcus aureus.
Microbiology (Reading): 2007, 153(Pt 12);4274-4283
[PubMed:18048940] [WorldCat.org] [DOI] (P p)
