Jump to navigation
Jump to search
NCBI: 03-AUG-2016
⊟Summary[edit | edit source]
- organism: Staphylococcus aureus NCTC8325
- locus tag: SAOUHSC_01253
- pan locus tag?: SAUPAN003577000
- symbol: SAOUHSC_01253
- pan gene symbol?: spoIIIE
- synonym:
- product: hypothetical protein
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
- type: CDS
- locus tag: SAOUHSC_01253
- symbol: SAOUHSC_01253
- product: hypothetical protein
- replicon: chromosome
- strand: +
- coordinates: 1206478..1208721
- length: 2244
- essential: no DEG other strains
⊟Accession numbers[edit | edit source]
- Gene ID: 3919984 NCBI
- RefSeq: YP_499786 NCBI
- BioCyc: G1I0R-1171 BioCyc
- MicrobesOnline: 1289700 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421
481
541
601
661
721
781
841
901
961
1021
1081
1141
1201
1261
1321
1381
1441
1501
1561
1621
1681
1741
1801
1861
1921
1981
2041
2101
2161
2221ATGGTGTTGGGTGTTTTCCAATTAGGAATAATAGGTCGTCTAATTGACAGCTTCTTTAAT
TATTTATTTGGGTACAGTAGATATTTAACATATATTTTAGTACTCTTAGCAACTGGTTTT
ATTACATACTCTAAACGTATTCCTAAAACTAGACGAACGGCTGGTTCGATTGTATTGCAA
ATTGCATTGCTATTTGTATCACAGTTAGTTTTTCATTTTAATAGTGGTATCAAAGCTGAA
AGAGAACCTGTACTTTCTTATGTGTATCAGTCATACCAACACAGTCATTTCCCAAATTTT
GGTGGCGGTGTATTAGGCTTTTATTTATTAGAGTTAAGCGTACCTTTAATTTCATTATTT
GGTGTATGTATTATTACTATTTTATTATTATGCTCAAGTGTTATTTTATTAACAAACCAT
CAACATCGTGAAGTTGCAAAAGTTGCACTGGAAAATATAAAAGCTTGGTTTGGTTCATTT
AATGAAAAAATGTCGGAAAGAAACCAAGAAAAACAATTGAAGCGTGAAGAAAAAGCAAGA
CTTAAAGAAGAACAAAAGGCACGTCAAAATGAACAGCCACAAATAAAAGATGTGAGTGAT
TTTACGGAAGTGCCTCAAGAAAGAGATATTCCAATTTATGGGCATACTGAAAATGAAAGT
AAAAGCCAGAGTCAACCAAGTCGAAAAAAACGAGTGTTTGATGCAGAGAATAGTTCGAAT
AACATCGTAAATCATCATCAAGCAGATCAGCAAGAACAATTAACAGAACAAACTCATAAC
AGTGTTGAAAGTGAAAACACTATTGAAGAAGCTGGTGAAGTTACGAATGTATCGTATGTT
GTTCCACCGTTAACTTTACTTAATCAACCTGCAAAACAAAAAGCAACATCTAAAGCTGAA
GTGCAACGTAAAGGACAAGTACTAGAGAATACATTAAAAGATTTTGGGGTAAATGCAAAA
GTGACACAAATTAAAATTGGTCCTGCAGTAACTCAATATGAAATTCAACCAGCTCAAGGG
GTTAAAGTGAGTAAAATTGTAAACTTGCATAATGATATTGCATTAGCTTTAGCAGCAAAA
GATGTTAGAATCGAAGCGCCAATACCTGGTCGTTCTGCAGTAGGTATTGAAGTGCCAAAT
GAGAAAATTTCATTAGTTTCACTAAAAGAAGTTTTAGATGAAAAATTCCCGTCTAATAAT
AAACTAGAAGTTGGATTAGGAAGAGATATATCAGGTGATCCAATTACTGTTCCACTAAAT
GAAATGCCACACTTATTGGTGGCAGGATCGACGGGTAGTGGTAAATCTGTTTGTATAAAT
GGTATTATTACAAGTATTTTATTAAATGCTAAGCCGCATGAAGTTAAACTTATGTTAATC
GATCCGAAAATGGTTGAACTAAATGTTTATAACGGAATTCCACACTTATTAATTCCGGTT
GTTACAAATCCTCATAAAGCTGCTCAAGCTTTAGAAAAAATTGTAGCTGAGATGGAAAGA
CGTTATGATTTATTCCAACATTCATCAACTAGAAACATTAAAGGTTATAACGAATTAATC
CGTAAGCAAAATCAAGAATTAGATGAGAAGCAACCAGAATTACCTTATATCGTTGTTATT
GTAGATGAGCTTGCAGATTTAATGATGGTAGCTGGTAAAGAAGTTGAAAATGCGATTCAA
CGTATTACACAAATGGCACGTGCAGCAGGTATACATTTAATTGTAGCGACACAAAGACCT
TCTGTGGATGTAATTACAGGTATCATTAAAAATAATATTCCATCTAGAATAGCTTTTGCT
GTGAGTTCTCAAACAGATTCAAGAACTATTATTGGTACTGGCGGCGCAGAAAAGTTACTT
GGTAAAGGTGACATGTTATACGTTGGAAATGGTGACTCATCACAAACACGTATTCAAGGG
GCGTTTTTAAGTGACCAAGAGGTGCAAGATGTTGTAAATTATGTAGTAGAACAACAACAG
GCAAATTATGTAAAAGAAATGGAACCAGATGCACCAGTGGATAAATCGGAAATGAAAAGT
GAAGATGCTTTATATGATGAAGCGTATTTGTTTGTTGTTGAACAACAAAAGGCAAGTACA
TCATTGTTACAACGCCAATTTAGAATTGGTTATAATAGAGCATCTAGGTTGATGGATGAT
TTAGAACGCAATCAGGTAATCGGTCCACAAAAAGGAAGCAAGCCTAGACAAGTTTTAATA
GATCTTAATAATGACGAGGTGTAA60
120
180
240
300
360
420
480
540
600
660
720
780
840
900
960
1020
1080
1140
1200
1260
1320
1380
1440
1500
1560
1620
1680
1740
1800
1860
1920
1980
2040
2100
2160
2220
2244
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SAOUHSC_01253
- symbol: SAOUHSC_01253
- description: hypothetical protein
- length: 747
- theoretical pI: 6.62784
- theoretical MW: 83404.8
- GRAVY: -0.264391
⊟Function[edit | edit source]
- TIGRFAM: Protein fate Protein and peptide secretion and trafficking type VII secretion protein EssC (TIGR03928; HMM-score: 124.1)Protein fate Protein and peptide secretion and trafficking type VII secretion protein EccCa (TIGR03924; HMM-score: 103)and 4 moreProtein fate Protein and peptide secretion and trafficking type VII secretion protein EccCb (TIGR03925; HMM-score: 65.2)Mobile and extrachromosomal element functions Other conjugative coupling factor TraD, SXT/TOL subfamily (TIGR03743; HMM-score: 15)type IV conjugative transfer system coupling protein TraD (TIGR02759; HMM-score: 12.4)Transport and binding proteins Cations and iron carrying compounds cation transporter family protein (TIGR00860; HMM-score: 11.3)
- TheSEED :
- Cell division protein FtsK
- PFAM: P-loop_NTPase (CL0023) FtsK_SpoIIIE; FtsK/SpoIIIE family (PF01580; HMM-score: 232.5)and 16 moreno clan defined FtsK_alpha; FtsK alpha domain (PF17854; HMM-score: 121.2)HTH (CL0123) FtsK_gamma; Ftsk gamma domain (PF09397; HMM-score: 99.9)no clan defined FtsK_4TM; 4TM region of DNA translocase FtsK/SpoIIIE (PF13491; HMM-score: 24.5)P-loop_NTPase (CL0023) AAA_10; AAA-like domain (PF12846; HMM-score: 21.6)AAA_19; AAA domain (PF13245; HMM-score: 20.1)DUF87; Helicase HerA, central domain (PF01935; HMM-score: 18.5)nSTAND1; Novel STAND NTPase 1 (PF20703; HMM-score: 17.3)ClpP_crotonase (CL0127) Peptidase_S49_N; Peptidase family S49 N-terminal (PF08496; HMM-score: 15.3)P-loop_NTPase (CL0023) AAA_22; AAA domain (PF13401; HMM-score: 14.8)no clan defined Neur_chan_memb; Neurotransmitter-gated ion-channel transmembrane region (PF02932; HMM-score: 13.9)LMBR1; LMBR1-like membrane protein (PF04791; HMM-score: 13.8)P-loop_NTPase (CL0023) AAA; ATPase family associated with various cellular activities (AAA) (PF00004; HMM-score: 13.4)AAA_29; P-loop containing region of AAA domain (PF13555; HMM-score: 12.6)ResIII; Type III restriction enzyme, res subunit (PF04851; HMM-score: 11.9)no clan defined Hikeshi-like_C; Hikeshi-like, C-terminal domain (PF21057; HMM-score: 11.7)Peptidase_AD (CL0130) Presenilin; Presenilin (PF01080; HMM-score: 6.2)
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors:
- effectors:
⊟Localization[edit | edit source]
- PSORTb: Cytoplasmic Membrane
- Cytoplasmic Score: 0
- Cytoplasmic Membrane Score: 10
- Cellwall Score: 0
- Extracellular Score: 0
- Internal Helices: 3
- DeepLocPro: Cytoplasmic Membrane
- Cytoplasmic Score: 0.0008
- Cytoplasmic Membrane Score: 0.9962
- Cell wall & surface Score: 0.0003
- Extracellular Score: 0.0027
- LocateP: Multi-transmembrane
- Prediction by SwissProt Classification: Membrane
- Pathway Prediction: Sec-(SPI)
- Intracellular possibility: 0.17
- Signal peptide possibility: -1
- N-terminally Anchored Score: 1
- Predicted Cleavage Site: No CleavageSite
- SignalP: no predicted signal peptide
- SP(Sec/SPI): 0.002552
- TAT(Tat/SPI): 0.000169
- LIPO(Sec/SPII): 0.003648
- predicted transmembrane helices (TMHMM): 4
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MVLGVFQLGIIGRLIDSFFNYLFGYSRYLTYILVLLATGFITYSKRIPKTRRTAGSIVLQIALLFVSQLVFHFNSGIKAEREPVLSYVYQSYQHSHFPNFGGGVLGFYLLELSVPLISLFGVCIITILLLCSSVILLTNHQHREVAKVALENIKAWFGSFNEKMSERNQEKQLKREEKARLKEEQKARQNEQPQIKDVSDFTEVPQERDIPIYGHTENESKSQSQPSRKKRVFDAENSSNNIVNHHQADQQEQLTEQTHNSVESENTIEEAGEVTNVSYVVPPLTLLNQPAKQKATSKAEVQRKGQVLENTLKDFGVNAKVTQIKIGPAVTQYEIQPAQGVKVSKIVNLHNDIALALAAKDVRIEAPIPGRSAVGIEVPNEKISLVSLKEVLDEKFPSNNKLEVGLGRDISGDPITVPLNEMPHLLVAGSTGSGKSVCINGIITSILLNAKPHEVKLMLIDPKMVELNVYNGIPHLLIPVVTNPHKAAQALEKIVAEMERRYDLFQHSSTRNIKGYNELIRKQNQELDEKQPELPYIVVIVDELADLMMVAGKEVENAIQRITQMARAAGIHLIVATQRPSVDVITGIIKNNIPSRIAFAVSSQTDSRTIIGTGGAEKLLGKGDMLYVGNGDSSQTRIQGAFLSDQEVQDVVNYVVEQQQANYVKEMEPDAPVDKSEMKSEDALYDEAYLFVVEQQKASTSLLQRQFRIGYNRASRLMDDLERNQVIGPQKGSKPRQVLIDLNNDEV
⊟Experimental data[edit | edit source]
- experimentally validated: PeptideAtlas [1] [2]
- protein localization: data available for COL
- quantitative data / protein copy number per cell:
- interaction partners:
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
- MicrobesOnline: SAOUHSC_01253 > SAOUHSC_01254 > SAOUHSC_01255 > SAOUHSC_01256 > SAOUHSC_01257 > SAOUHSC_01258 > SAOUHSC_01259 > SAOUHSC_01260predicted SigA promoter [3] : S521 > SAOUHSC_01252 > S522 > SAOUHSC_01253 > SAOUHSC_01254 > SAOUHSC_01255 > SAOUHSC_01256 > SAOUHSC_01257 > S523 > SAOUHSC_01258 > SAOUHSC_01259 > SAOUHSC_01260
⊟Regulation[edit | edit source]
- regulator:
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: [3]
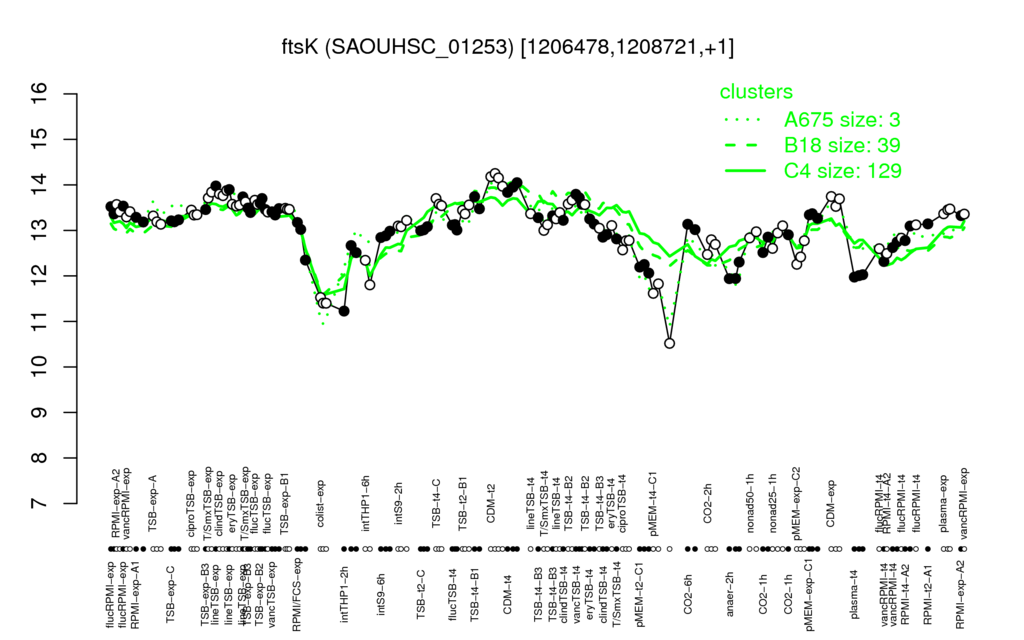 Multi-gene expression profiles
Multi-gene expression profiles
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
- Aureolib: no data available
⊟Protein stability[edit | edit source]
- half-life: no data available
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You can add further information about the gene and protein here. [edit]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ Maren Depke, Stephan Michalik, Alexander Rabe, Kristin Surmann, Lars Brinkmann, Nico Jehmlich, Jörg Bernhardt, Michael Hecker, Bernd Wollscheid, Zhi Sun, Robert L Moritz, Uwe Völker, Frank Schmidt
A peptide resource for the analysis of Staphylococcus aureus in host-pathogen interaction studies.
Proteomics: 2015, 15(21);3648-61
[PubMed:26224020] [WorldCat.org] [DOI] (I p) - ↑ Stephan Michalik, Maren Depke, Annette Murr, Manuela Gesell Salazar, Ulrike Kusebauch, Zhi Sun, Tanja C Meyer, Kristin Surmann, Henrike Pförtner, Petra Hildebrandt, Stefan Weiss, Laura Marcela Palma Medina, Melanie Gutjahr, Elke Hammer, Dörte Becher, Thomas Pribyl, Sven Hammerschmidt, Eric W Deutsch, Samuel L Bader, Michael Hecker, Robert L Moritz, Ulrike Mäder, Uwe Völker, Frank Schmidt
A global Staphylococcus aureus proteome resource applied to the in vivo characterization of host-pathogen interactions.
Sci Rep: 2017, 7(1);9718
[PubMed:28887440] [WorldCat.org] [DOI] (I e) - ↑ 3.0 3.1 Ulrike Mäder, Pierre Nicolas, Maren Depke, Jan Pané-Farré, Michel Debarbouille, Magdalena M van der Kooi-Pol, Cyprien Guérin, Sandra Dérozier, Aurelia Hiron, Hanne Jarmer, Aurélie Leduc, Stephan Michalik, Ewoud Reilman, Marc Schaffer, Frank Schmidt, Philippe Bessières, Philippe Noirot, Michael Hecker, Tarek Msadek, Uwe Völker, Jan Maarten van Dijl
Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions.
PLoS Genet: 2016, 12(4);e1005962
[PubMed:27035918] [WorldCat.org] [DOI] (I e)
