⊟Summary[edit | edit source]
- organism: Staphylococcus aureus NCTC8325
- locus tag: SAOUHSC_02706
- pan locus tag?: SAUPAN005955000
- symbol: SAOUHSC_02706
- pan gene symbol?: sbi
- synonym:
- product: immunoglobulin G-binding protein Sbi
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
- type: CDS
- locus tag: SAOUHSC_02706
- symbol: SAOUHSC_02706
- product: immunoglobulin G-binding protein Sbi
- replicon: chromosome
- strand: +
- coordinates: 2487383..2488693
- length: 1311
- essential: no DEG other strains
⊟Accession numbers[edit | edit source]
- Gene ID: 3919725 NCBI
- RefSeq: YP_501168 NCBI
- BioCyc: G1I0R-2552 BioCyc
- MicrobesOnline: 1291139 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421
481
541
601
661
721
781
841
901
961
1021
1081
1141
1201
1261ATGAAAAATAAATATATCTCGAAGTTGCTAGTTGGGGCAGCAACAATTACGTTAGCTACA
ATGATTTCAAATGGGGAAGCAAAAGCGAGTGAAAACACGCAACAAACTTCAACTAAGCAC
CAAACAACTCAAAACAACTACGTAACAGATCAACAAAAAGCTTTTTATCAAGTATTACAT
CTAAAAGGTATCACAGAAGAACAACGTAACCAATACATCAAAACATTACGCGAACACCCA
GAACGTGCACAAGAAGTATTCTCTGAATCACTTAAAGACAGCAAGAACCCAGACCGACGT
GTTGCACAACAAAACGCTTTTTACAATGTTCTTAAAAATGATAACTTAACTGAACAAGAA
AAAAATAATTACATTGCACAAATTAAAGAAAACCCTGATAGAAGCCAACAAGTTTGGGTA
GAATCAGTACAATCTTCTAAAGCTAAAGAACGTCAAAATATTGAAAATGCGGATAAAGCA
ATTAAAGATTTCCAAGATAACAAAGCACCACACGATAAATCAGCAGCATATGAAGCTAAC
TCAAAATTACCTAAAGATTTACGTGATAAAAACAACCGCTTTGTAGAAAAAGTTTCAATT
GAAAAAGCAATCGTTCGTCATGATGAGCGTGTGAAATCAGCAAATGATGCAATCTCAAAA
TTAAATGAAAAAGATTCAATTGAAAACAGACGTTTAGCACAACGTGAAGTTAACAAAGCA
CCTATGGATGTAAAAGAGCATTTACAGAAACAATTAGACGCATTAGTTGCTCAAAAAGAT
GCTGAAAAGAAAGTGGCGCCAAAAGTTGAGGCTCCTCAAATTCAATCACCACAAATTGAA
AAACCTAAAGTAGAATCACCAAAAGTTGAAGTCCCTCAAATTCAATCACCAAAAGTTGAG
GTTCCTCAATCTAAATTATTAGGTTACTACCAATCATTAAAAGATTCATTTAACTATGGT
TACAAGTATTTAACAGATACTTATAAAAGCTATAAAGAAAAATATGATACAGCAAAGTAC
TACTATAATACGTACTATAAATACAAAGGTGCGATTGATCAAACAGTATTAACAGTACTA
GGTAGTGGTTCTAAATCTTACATCCAACCATTGAAAGTTGATGATAAAAACGGCTACTTA
GCTAAATCATATGCACAAGTAAGAAACTATGTAACTGAGTCAATCAATACTGGTAAAGTA
TTATATACTTTCTACCAAAACCCAACATTAGTAAAAACAGCTATTAAAGCTCAAGAAACT
GCATCATCAATCAAAAATACATTAAGTAATTTATTATCATTCTGGAAATAA60
120
180
240
300
360
420
480
540
600
660
720
780
840
900
960
1020
1080
1140
1200
1260
1311
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SAOUHSC_02706
- symbol: SAOUHSC_02706
- description: immunoglobulin G-binding protein Sbi
- length: 436
- theoretical pI: 9.87462
- theoretical MW: 50069.9
- GRAVY: -0.961009
⊟Function[edit | edit source]
- TIGRFAM:
- TheSEED :
- IgG-binding protein SBI
- PFAM: B_GA (CL0598) B; B domain (PF02216; HMM-score: 178.4)no clan defined Sbi-IV; C3 binding domain 4 of IgG-bind protein SBI (PF11621; HMM-score: 155.5)and 3 moreDUF5392; Family of unknown function (DUF5392) (PF17370; HMM-score: 21)Extensin_1; Extensin-like protein (PF02095; HMM-score: 15.2)DUF1539; Domain of Unknown Function (DUF1539) (PF07560; HMM-score: 13.7)
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors:
- effectors:
⊟Localization[edit | edit source]
- PSORTb: unknown (no significant prediction)
- Cytoplasmic Score: 0
- Cytoplasmic Membrane Score: 3.33
- Cellwall Score: 3.33
- Extracellular Score: 3.33
- Internal Helices: 0
- DeepLocPro: Extracellular
- Cytoplasmic Score: 0
- Cytoplasmic Membrane Score: 0.001
- Cell wall & surface Score: 0.0408
- Extracellular Score: 0.9582
- LocateP: Secretory(released) (with CS)
- Prediction by SwissProt Classification: Extracellular
- Pathway Prediction: Sec-(SPI)
- Intracellular possibility: -0.17
- Signal peptide possibility: 1
- N-terminally Anchored Score: 0
- Predicted Cleavage Site: GEAKASEN
- SignalP: Signal peptide SP(Sec/SPI) length 29 aa
- SP(Sec/SPI): 0.989464
- TAT(Tat/SPI): 0.00375
- LIPO(Sec/SPII): 0.005096
- Cleavage Site: CS pos: 29-30. AKA-SE. Pr: 0.9454
- predicted transmembrane helices (TMHMM): 0
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MKNKYISKLLVGAATITLATMISNGEAKASENTQQTSTKHQTTQNNYVTDQQKAFYQVLHLKGITEEQRNQYIKTLREHPERAQEVFSESLKDSKNPDRRVAQQNAFYNVLKNDNLTEQEKNNYIAQIKENPDRSQQVWVESVQSSKAKERQNIENADKAIKDFQDNKAPHDKSAAYEANSKLPKDLRDKNNRFVEKVSIEKAIVRHDERVKSANDAISKLNEKDSIENRRLAQREVNKAPMDVKEHLQKQLDALVAQKDAEKKVAPKVEAPQIQSPQIEKPKVESPKVEVPQIQSPKVEVPQSKLLGYYQSLKDSFNYGYKYLTDTYKSYKEKYDTAKYYYNTYYKYKGAIDQTVLTVLGSGSKSYIQPLKVDDKNGYLAKSYAQVRNYVTESINTGKVLYTFYQNPTLVKTAIKAQETASSIKNTLSNLLSFWK
⊟Experimental data[edit | edit source]
- experimentally validated: PeptideAtlas [1] [2]
- protein localization: data available for COL
- quantitative data / protein copy number per cell:
- interaction partners:
SAOUHSC_01719 hypothetical protein [3] (data from MRSA252) SAOUHSC_02366 fructose-bisphosphate aldolase [3] (data from MRSA252)
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
- predicted SigA promoter [4] : SAOUHSC_02706 > S1054
⊟Regulation[edit | edit source]
- regulators: SaeR* (activation) regulon, RNAIII/S871 regulon
SaeR* (TF) important in Virulence; RegPrecise RNAIII/S871 (sRNA) important in Virulence, Quorum sensing; [5]
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: [4]
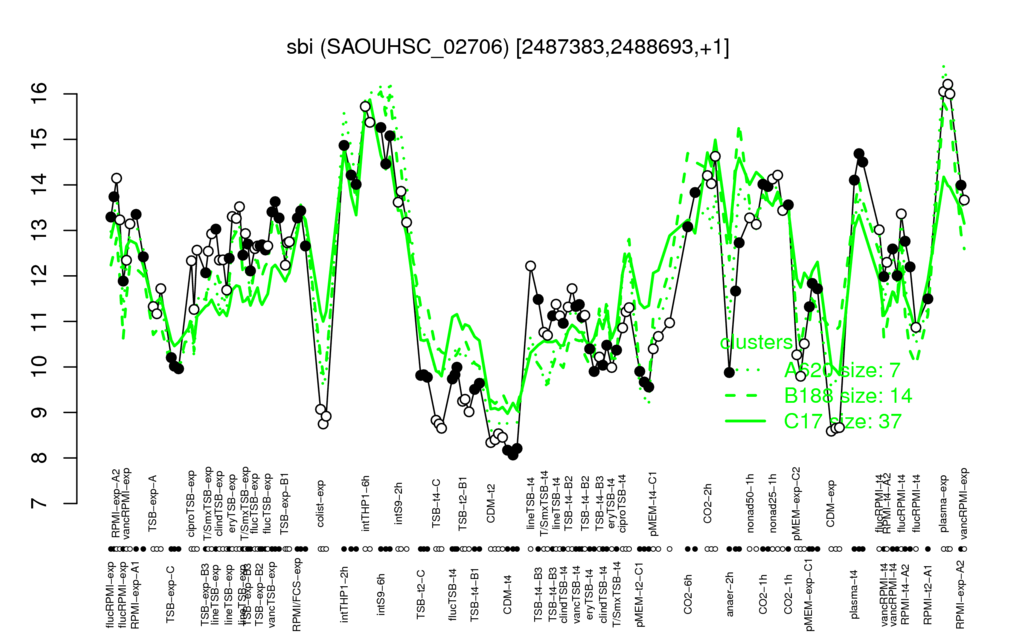 Multi-gene expression profiles
Multi-gene expression profiles
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
- Aureolib: no data available
⊟Protein stability[edit | edit source]
- half-life: no data available
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You can add further information about the gene and protein here. [edit]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ Maren Depke, Stephan Michalik, Alexander Rabe, Kristin Surmann, Lars Brinkmann, Nico Jehmlich, Jörg Bernhardt, Michael Hecker, Bernd Wollscheid, Zhi Sun, Robert L Moritz, Uwe Völker, Frank Schmidt
A peptide resource for the analysis of Staphylococcus aureus in host-pathogen interaction studies.
Proteomics: 2015, 15(21);3648-61
[PubMed:26224020] [WorldCat.org] [DOI] (I p) - ↑ Stephan Michalik, Maren Depke, Annette Murr, Manuela Gesell Salazar, Ulrike Kusebauch, Zhi Sun, Tanja C Meyer, Kristin Surmann, Henrike Pförtner, Petra Hildebrandt, Stefan Weiss, Laura Marcela Palma Medina, Melanie Gutjahr, Elke Hammer, Dörte Becher, Thomas Pribyl, Sven Hammerschmidt, Eric W Deutsch, Samuel L Bader, Michael Hecker, Robert L Moritz, Ulrike Mäder, Uwe Völker, Frank Schmidt
A global Staphylococcus aureus proteome resource applied to the in vivo characterization of host-pathogen interactions.
Sci Rep: 2017, 7(1);9718
[PubMed:28887440] [WorldCat.org] [DOI] (I e) - ↑ 3.0 3.1 Artem Cherkasov, Michael Hsing, Roya Zoraghi, Leonard J Foster, Raymond H See, Nikolay Stoynov, Jihong Jiang, Sukhbir Kaur, Tian Lian, Linda Jackson, Huansheng Gong, Rick Swayze, Emily Amandoron, Farhad Hormozdiari, Phuong Dao, Cenk Sahinalp, Osvaldo Santos-Filho, Peter Axerio-Cilies, Kendall Byler, William R McMaster, Robert C Brunham, B Brett Finlay, Neil E Reiner
Mapping the protein interaction network in methicillin-resistant Staphylococcus aureus.
J Proteome Res: 2011, 10(3);1139-50
[PubMed:21166474] [WorldCat.org] [DOI] (I p) - ↑ 4.0 4.1 Ulrike Mäder, Pierre Nicolas, Maren Depke, Jan Pané-Farré, Michel Debarbouille, Magdalena M van der Kooi-Pol, Cyprien Guérin, Sandra Dérozier, Aurelia Hiron, Hanne Jarmer, Aurélie Leduc, Stephan Michalik, Ewoud Reilman, Marc Schaffer, Frank Schmidt, Philippe Bessières, Philippe Noirot, Michael Hecker, Tarek Msadek, Uwe Völker, Jan Maarten van Dijl
Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions.
PLoS Genet: 2016, 12(4);e1005962
[PubMed:27035918] [WorldCat.org] [DOI] (I e) - ↑ Delphine Bronesky, Zongfu Wu, Stefano Marzi, Philippe Walter, Thomas Geissmann, Karen Moreau, François Vandenesch, Isabelle Caldelari, Pascale Romby
Staphylococcus aureus RNAIII and Its Regulon Link Quorum Sensing, Stress Responses, Metabolic Adaptation, and Regulation of Virulence Gene Expression.
Annu Rev Microbiol: 2016, 70;299-316
[PubMed:27482744] [WorldCat.org] [DOI] (I p)
⊟Relevant publications[edit | edit source]
Lihong Zhang, Karin Jacobsson, Katrin Ström, Martin Lindberg, Lars Frykberg
Staphylococcus aureus expresses a cell surface protein that binds both IgG and beta2-glycoprotein I.
Microbiology (Reading): 1999, 145 ( Pt 1);177-183
[PubMed:10206697] [WorldCat.org] [DOI] (P p)L Zhang, A Rosander, K Jacobsson, M Lindberg, L Frykberg
Expression of staphylococcal protein Sbi is induced by human IgG.
FEMS Immunol Med Microbiol: 2000, 28(3);211-8
[PubMed:10865173] [WorldCat.org] [DOI] (P p)Mark J J B Sibbald, Theresa Winter, Magdalena M van der Kooi-Pol, G Buist, E Tsompanidou, Tjibbe Bosma, Tina Schäfer, Knut Ohlsen, Michael Hecker, Haike Antelmann, Susanne Engelmann, Jan Maarten van Dijl
Synthetic effects of secG and secY2 mutations on exoproteome biogenesis in Staphylococcus aureus.
J Bacteriol: 2010, 192(14);3788-800
[PubMed:20472795] [WorldCat.org] [DOI] (I p)Hwan Keun Kim, Vilasack Thammavongsa, Olaf Schneewind, Dominique Missiakas
Recurrent infections and immune evasion strategies of Staphylococcus aureus.
Curr Opin Microbiol: 2012, 15(1);92-9
[PubMed:22088393] [WorldCat.org] [DOI] (I p)K Jacobsson, L Frykberg
Cloning of ligand-binding domains of bacterial receptors by phage display.
Biotechniques: 1995, 18(5);878-85
[PubMed:7619494] [WorldCat.org] (P p)L Zhang, K Jacobsson, J Vasi, M Lindberg, L Frykberg
A second IgG-binding protein in Staphylococcus aureus.
Microbiology (Reading): 1998, 144 ( Pt 4);985-91
[PubMed:9579072] [WorldCat.org] [DOI] (P p)
