Jump to navigation
Jump to search
NCBI: 10-JUN-2013
⊟Summary[edit | edit source]
- organism: Staphylococcus aureus COL
- locus tag: SACOL2105 [new locus tag: SACOL_RS11015 ]
- pan locus tag?: SAUPAN005404000
- symbol: glyA
- pan gene symbol?: glyA
- synonym:
- product: serine hydroxymethyltransferase
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
- type: CDS
- locus tag: SACOL2105 [new locus tag: SACOL_RS11015 ]
- symbol: glyA
- product: serine hydroxymethyltransferase
- replicon: chromosome
- strand: -
- coordinates: 2165590..2166828
- length: 1239
- essential: unknown other strains
⊟Accession numbers[edit | edit source]
- Gene ID: 3237486 NCBI
- RefSeq: YP_186920 NCBI
- BioCyc: see SACOL_RS11015
- MicrobesOnline: 913581 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421
481
541
601
661
721
781
841
901
961
1021
1081
1141
1201ATGTCTTATATCACCAAGCAAGATAAAGTTATCGCAGAAGCAATCGAGAGAGAATTTCAG
AGACAAAATAGCAACATAGAGTTAATCGCATCGGAAAATTTTGTATCGGAAGCGGTTATG
GAAGCACAAGGTTCAGTGTTGACTAATAAGTATGCTGAAGGCTATCCAGGACGCCGATAT
TATGGTGGCTGTGAGTTTGTAGATGTTACTGAAAGCATCGCAATTGATCGTGCTAAAGCA
TTGTTTGGAGCTGAACATGTCAATGTTCAACCACATTCAGGTTCACAAGCGAACATGGCT
GTTTACTTAGTTGCATTAGAAATGGGCGACACAGTTTTAGGTATGAATTTGAGTCATGGT
GGTCACTTGACACATGGAGCGCCTGTTAATTTTAGTGGTAAATTCTACAATTTCGTTGAA
TATGGAGTAGATAAAGACACAGAACGAATCAATTATGATGAAGTTCGTAAATTAGCGTTA
GAGCATAAGCCTAAGCTTATTGTGGCAGGAGCATCAGCATATTCAAGAACAATTGACTTC
AAAAAGTTTAAAGAAATCGCAGATGAAGTAAACGCTAAGTTAATGGTAGACATGGCACAT
ATTGCAGGATTAGTAGCGGCAGGTTTACATCCAAATCCAGTAGAATATGCTGATTTTGTA
ACAACTACAACACACAAAACATTACGCGGACCACGTGGTGGTATGATTTTATGTAAGGAA
GAATATAAAAAAGACATAGATAAAACAATTTTCCCTGGTATTCAAGGTGGACCTCTTGAG
CATGTTATTGCAGCAAAAGCAGTTGCTTTTGGAGAAGCGTTAGAAAATAATTTCAAAACG
TATCAACAACAAGTGGTTAAAAACGCAAAAGTTCTTGCAGAAGCATTAATTAATGAAGGA
TTTAGAATTGTTTCTGGCGGTACAGATAATCACTTAGTAGCTGTTGATGTAAAAGGGTCT
ATAGGACTTACTGGTAAAGAAGCTGAAGAGACTTTAGATTCAGTTGGTATCACATGTAAC
AAAAATACCATTCCGTTCGATCAAGAAAAACCTTTTGTAACGAGTGGTATACGTTTAGGT
ACACCTGCTGCAACAACGCGTGGATTTGATGAAAAAGCTTTTGAGGAAGTTGCAAAAATC
ATCAGTTTAGCATTGAAAAATAGTAAAGATGAAGAAAAATTACAACAAGCTAAAGAACGC
GTTGCGAAATTAACAGCTGAATATCCTCTATATCAATAA60
120
180
240
300
360
420
480
540
600
660
720
780
840
900
960
1020
1080
1140
1200
1239
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SACOL2105 [new locus tag: SACOL_RS11015 ]
- symbol: GlyA
- description: serine hydroxymethyltransferase
- length: 412
- theoretical pI: 5.99288
- theoretical MW: 45172
- GRAVY: -0.269417
⊟Function[edit | edit source]
- reaction: EC 2.1.2.1? ExPASyGlycine hydroxymethyltransferase 5,10-methylenetetrahydrofolate + glycine + H2O = tetrahydrofolate + L-serine
- TIGRFAM: Cellular processes Biosynthesis of natural products fluorothreonine transaldolase (TIGR04506; EC 2.2.1.8; HMM-score: 114.6)and 7 moreBiosynthesis of cofactors, prosthetic groups, and carriers Biotin 8-amino-7-oxononanoate synthase (TIGR00858; EC 2.3.1.47; HMM-score: 26.8)Unknown function Enzymes of unknown specificity cysteine desulfurase family protein (TIGR01977; HMM-score: 22.3)Unknown function Enzymes of unknown specificity pyridoxal phosphate enzyme, MJ0158 family (TIGR03576; HMM-score: 20)Energy metabolism Amino acids and amines glycine C-acetyltransferase (TIGR01822; EC 2.3.1.29; HMM-score: 18.8)UDP-4-amino-4,6-dideoxy-N-acetyl-beta-L-altrosamine transaminase (TIGR03588; EC 2.6.1.92; HMM-score: 16.6)dTDP-4-dehydro-6-deoxyglucose aminotransferase (TIGR04427; EC 2.-.-.-; HMM-score: 14.1)putative methanogenesis marker protein 8 (TIGR03275; HMM-score: 12.8)
- TheSEED :
- Serine hydroxymethyltransferase (EC 2.1.2.1)
Amino Acids and Derivatives Alanine, serine, and glycine Glycine and Serine Utilization Serine hydroxymethyltransferase (EC 2.1.2.1)and 6 moreAmino Acids and Derivatives Alanine, serine, and glycine Glycine Biosynthesis Serine hydroxymethyltransferase (EC 2.1.2.1)Amino Acids and Derivatives Alanine, serine, and glycine Glycine cleavage system Serine hydroxymethyltransferase (EC 2.1.2.1)Amino Acids and Derivatives Alanine, serine, and glycine Serine Biosynthesis Serine hydroxymethyltransferase (EC 2.1.2.1)Carbohydrates One-carbon Metabolism Serine-glyoxylate cycle Serine hydroxymethyltransferase (EC 2.1.2.1) - PFAM: PLP_aminotran (CL0061) SHMT; Serine hydroxymethyltransferase (PF00464; HMM-score: 589.1)and 5 moreBeta_elim_lyase; Beta-eliminating lyase (PF01212; HMM-score: 29.9)Aminotran_1_2; Aminotransferase class I and II (PF00155; HMM-score: 28)DegT_DnrJ_EryC1; DegT/DnrJ/EryC1/StrS aminotransferase family (PF01041; HMM-score: 13.4)Aminotran_5; Aminotransferase class-V (PF00266; HMM-score: 13.1)Cys_Met_Meta_PP; Cys/Met metabolism PLP-dependent enzyme (PF01053; HMM-score: 11.5)
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors: pyridoxal 5'-phosphate
- effectors:
⊟Localization[edit | edit source]
- PSORTb: Cytoplasmic
- Cytoplasmic Score: 9.97
- Cytoplasmic Membrane Score: 0
- Cellwall Score: 0.01
- Extracellular Score: 0.02
- Internal Helices: 0
- DeepLocPro: Cytoplasmic
- Cytoplasmic Score: 0.8918
- Cytoplasmic Membrane Score: 0.0012
- Cell wall & surface Score: 0.0001
- Extracellular Score: 0.1069
- LocateP: Intracellular
- Prediction by SwissProt Classification: Cytoplasmic
- Pathway Prediction: No pathway
- Intracellular possibility: 1
- Signal peptide possibility: -1
- N-terminally Anchored Score: -1
- Predicted Cleavage Site: No CleavageSite
- SignalP: no predicted signal peptide
- SP(Sec/SPI): 0.005746
- TAT(Tat/SPI): 0.001573
- LIPO(Sec/SPII): 0.000815
- predicted transmembrane helices (TMHMM): 0
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MSYITKQDKVIAEAIEREFQRQNSNIELIASENFVSEAVMEAQGSVLTNKYAEGYPGRRYYGGCEFVDVTESIAIDRAKALFGAEHVNVQPHSGSQANMAVYLVALEMGDTVLGMNLSHGGHLTHGAPVNFSGKFYNFVEYGVDKDTERINYDEVRKLALEHKPKLIVAGASAYSRTIDFKKFKEIADEVNAKLMVDMAHIAGLVAAGLHPNPVEYADFVTTTTHKTLRGPRGGMILCKEEYKKDIDKTIFPGIQGGPLEHVIAAKAVAFGEALENNFKTYQQQVVKNAKVLAEALINEGFRIVSGGTDNHLVAVDVKGSIGLTGKEAEETLDSVGITCNKNTIPFDQEKPFVTSGIRLGTPAATTRGFDEKAFEEVAKIISLALKNSKDEEKLQQAKERVAKLTAEYPLYQ
⊟Experimental data[edit | edit source]
- experimentally validated: PeptideAtlas
- protein localization: Cytoplasmic [1] [2] [3] [4]
- quantitative data / protein copy number per cell: 2758 [5]
- interaction partners:
SACOL1102 (pdhA) pyruvate dehydrogenase complex E1 component subunit alpha [6] (data from MRSA252) SACOL2227 (rplE) 50S ribosomal protein L5 [6] (data from MRSA252) SACOL2222 (rpsE) 30S ribosomal protein S5 [6] (data from MRSA252) SACOL0594 (tuf) elongation factor Tu [6] (data from MRSA252) SACOL2104 (upp) uracil phosphoribosyltransferase [6] (data from MRSA252) SACOL0211 acetyl-CoA acetyltransferase [6] (data from MRSA252)
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
- MicrobesOnline: SACOL2103 < upp < glyA < SACOL2106
⊟Regulation[edit | edit source]
- regulator:
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: data available for NCTC8325
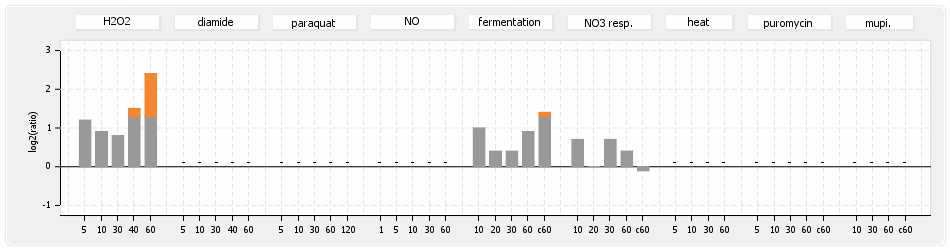
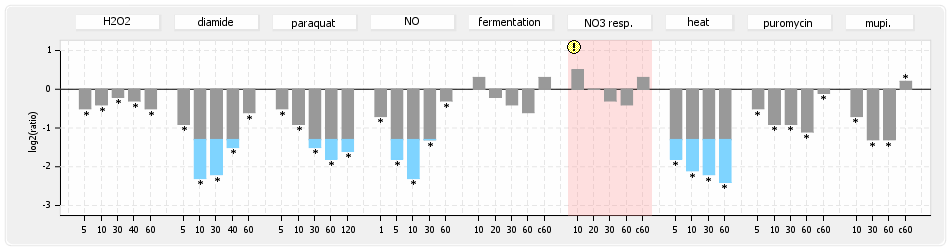
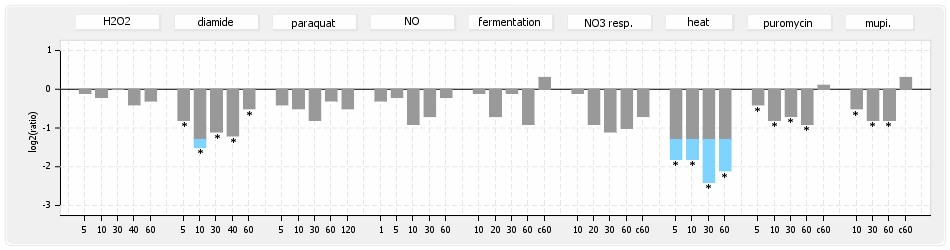
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
⊟Protein stability[edit | edit source]
- half-life: 19.22 h [7]
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You can add further information about the gene and protein here. [edit]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ Dörte Becher, Kristina Hempel, Susanne Sievers, Daniela Zühlke, Jan Pané-Farré, Andreas Otto, Stephan Fuchs, Dirk Albrecht, Jörg Bernhardt, Susanne Engelmann, Uwe Völker, Jan Maarten van Dijl, Michael Hecker
A proteomic view of an important human pathogen--towards the quantification of the entire Staphylococcus aureus proteome.
PLoS One: 2009, 4(12);e8176
[PubMed:19997597] [WorldCat.org] [DOI] (I e) - ↑ Kristina Hempel, Jan Pané-Farré, Andreas Otto, Susanne Sievers, Michael Hecker, Dörte Becher
Quantitative cell surface proteome profiling for SigB-dependent protein expression in the human pathogen Staphylococcus aureus via biotinylation approach.
J Proteome Res: 2010, 9(3);1579-90
[PubMed:20108986] [WorldCat.org] [DOI] (I p) - ↑ Kristina Hempel, Florian-Alexander Herbst, Martin Moche, Michael Hecker, Dörte Becher
Quantitative proteomic view on secreted, cell surface-associated, and cytoplasmic proteins of the methicillin-resistant human pathogen Staphylococcus aureus under iron-limited conditions.
J Proteome Res: 2011, 10(4);1657-66
[PubMed:21323324] [WorldCat.org] [DOI] (I p) - ↑ Andreas Otto, Jan Maarten van Dijl, Michael Hecker, Dörte Becher
The Staphylococcus aureus proteome.
Int J Med Microbiol: 2014, 304(2);110-20
[PubMed:24439828] [WorldCat.org] [DOI] (I p) - ↑ Daniela Zühlke, Kirsten Dörries, Jörg Bernhardt, Sandra Maaß, Jan Muntel, Volkmar Liebscher, Jan Pané-Farré, Katharina Riedel, Michael Lalk, Uwe Völker, Susanne Engelmann, Dörte Becher, Stephan Fuchs, Michael Hecker
Costs of life - Dynamics of the protein inventory of Staphylococcus aureus during anaerobiosis.
Sci Rep: 2016, 6;28172
[PubMed:27344979] [WorldCat.org] [DOI] (I e) - ↑ 6.0 6.1 6.2 6.3 6.4 6.5 Artem Cherkasov, Michael Hsing, Roya Zoraghi, Leonard J Foster, Raymond H See, Nikolay Stoynov, Jihong Jiang, Sukhbir Kaur, Tian Lian, Linda Jackson, Huansheng Gong, Rick Swayze, Emily Amandoron, Farhad Hormozdiari, Phuong Dao, Cenk Sahinalp, Osvaldo Santos-Filho, Peter Axerio-Cilies, Kendall Byler, William R McMaster, Robert C Brunham, B Brett Finlay, Neil E Reiner
Mapping the protein interaction network in methicillin-resistant Staphylococcus aureus.
J Proteome Res: 2011, 10(3);1139-50
[PubMed:21166474] [WorldCat.org] [DOI] (I p) - ↑ Stephan Michalik, Jörg Bernhardt, Andreas Otto, Martin Moche, Dörte Becher, Hanna Meyer, Michael Lalk, Claudia Schurmann, Rabea Schlüter, Holger Kock, Ulf Gerth, Michael Hecker
Life and death of proteins: a case study of glucose-starved Staphylococcus aureus.
Mol Cell Proteomics: 2012, 11(9);558-70
[PubMed:22556279] [WorldCat.org] [DOI] (I p)