Jump to navigation
Jump to search
NCBI: 03-AUG-2016
⊟Summary[edit | edit source]
- organism: Staphylococcus aureus NCTC8325
- locus tag: SAOUHSC_00849
- pan locus tag?: SAUPAN002897000
- symbol: SAOUHSC_00849
- pan gene symbol?: sufS
- synonym:
- product: aminotransferase
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
⊟Accession numbers[edit | edit source]
- Gene ID: 3918980 NCBI
- RefSeq: YP_499403 NCBI
- BioCyc: G1I0R-793 BioCyc
- MicrobesOnline: 1289314 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421
481
541
601
661
721
781
841
901
961
1021
1081
1141
1201GTGAATGAAGTGGCCGAACACTCATTTGACGTTAATGAAGTAATCAAGGATTTTCCGATA
TTAGATCAAAAAGTCAATGGCAAACGTTTAGCATATCTTGATTCAACAGCGACAAGTCAA
ACGCCTGTGCAAGTGTTAAATGTTTTAGAAGATTACTACAAGCGTTATAATTCAAACGTT
CATCGTGGTGTTCATACATTAGGATCATTGGCAACTGATGGTTATGAAAATGCCCGTGAA
ACCGTTCGTCGTTTTATTAATGCGAAGTATTTTGAAGAAATCATTTTTACACGCGGAACA
ACTGCGTCGATTAACCTTGTAGCACATAGCTATGGTGATGCAAATGTTGAAGAGGGCGAT
GAAATTGTTGTCACTGAAATGGAACATCATGCCAATATTGTTCCTTGGCAACAGTTAGCA
AAGCGTAAAAATGCGACATTGAAATTTATACCAATGACAGCTGACGGTGAATTAAACATC
GAGGATATTAAGCAAACGATTAATGATAAAACAAAGATCGTTGCTATTGCACATATATCT
AATGTGCTCGGTACAATTAATGATGTTAAAACCATTGCAGAAATAGCTCATCAACATGGT
GCAATTATCAGTGTTGATGGGGCGCAAGCAGCACCACATATGAAACTTGATATGCAAGAA
ATGAATGCTGATTTTTATAGTTTTAGTGGTCATAAAATGCTTGGACCAACAGGTATTGGC
GTATTATTTGGTAAACGTGAGTTACTACAAAAAATGGAACCGATTGAGTTCGGTGGCGAC
ATGATTGATTTTGTAAGTAAGTATGATGCAACATGGGCTGATTTACCTACTAAATTTGAG
GCGGGTACTCCATTAATTGCTCAAGCAATTGGGCTTGCAGAAGCTATTCGCTATTTAGAA
CGCATAGGTTTTGATGCAATTCATAAATATGAACAAGAATTAACGATATATGCTTATGAG
CAAATGTCTGCAATTGAAGGAATTGAAATTTATGGCCCGCCAAAGGATCGTCGTGCAGGT
GTAATAACGTTTAATTTACAAGATGTACATCCACACGATGTTGCTACAGCCGTAGATACA
GAAGGTGTAGCGGTTAGAGCTGGGCATCATTGTGCGCAACCGTTAATGAAATGGTTAAAT
GTGTCTTCAACAGCTAGAGCGAGTTTTTATATATACAACACGAAAGAAGACGTTGATCAG
TTAATAAATGCCTTGAAACAAACGAAGGAGTTTTTCTCTTATGAATTTTAA60
120
180
240
300
360
420
480
540
600
660
720
780
840
900
960
1020
1080
1140
1200
1251
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SAOUHSC_00849
- symbol: SAOUHSC_00849
- description: aminotransferase
- length: 416
- theoretical pI: 5.18484
- theoretical MW: 46618.4
- GRAVY: -0.257933
⊟Function[edit | edit source]
- reaction: EC 2.8.1.7? ExPASyCysteine desulfurase L-cysteine + acceptor = L-alanine + S-sulfanyl-acceptor
- TIGRFAM: Biosynthesis of cofactors, prosthetic groups, and carriers Other cysteine desulfurase, SufS family (TIGR01979; HMM-score: 614.9)and 33 moreBiosynthesis of cofactors, prosthetic groups, and carriers Other cysteine desulfurase, catalytic subunit CsdA (TIGR03392; EC 2.8.1.7; HMM-score: 385.1)Unknown function Enzymes of unknown specificity cysteine desulfurase family protein (TIGR01976; HMM-score: 260.8)Unknown function Enzymes of unknown specificity cysteine desulfurase family protein (TIGR01977; HMM-score: 251)cysteine desulfurase NifS (TIGR03402; EC 2.8.1.7; HMM-score: 167.4)DNA metabolism Restriction/modification cysteine desulfurase DndA (TIGR03235; EC 2.8.1.7; HMM-score: 144.7)Protein synthesis tRNA and rRNA base modification cysteine desulfurase IscS (TIGR02006; EC 2.8.1.7; HMM-score: 135.9)Biosynthesis of cofactors, prosthetic groups, and carriers Other cysteine desulfurase IscS (TIGR02006; EC 2.8.1.7; HMM-score: 135.9)cysteine desulfurase, NifS family (TIGR03403; EC 2.8.1.7; HMM-score: 119.5)Biosynthesis of cofactors, prosthetic groups, and carriers Glutathione and analogs ergothioneine biosynthesis PLP-dependent enzyme EgtE (TIGR04343; EC 4.4.-.-; HMM-score: 70.6)2-aminoethylphosphonate aminotransferase (TIGR03301; EC 2.6.1.-; HMM-score: 42.9)O-phospho-L-seryl-tRNA:Cys-tRNA synthase (TIGR02539; EC 2.5.1.73; HMM-score: 41.7)Energy metabolism Amino acids and amines kynureninase (TIGR01814; EC 3.7.1.3; HMM-score: 33.3)Amino acid biosynthesis Aspartate family O-succinylhomoserine sulfhydrylase (TIGR01325; EC 4.2.99.-; HMM-score: 33.1)Energy metabolism Amino acids and amines methionine gamma-lyase (TIGR01328; EC 4.4.1.11; HMM-score: 33)Central intermediary metabolism Phosphorus compounds 2-aminoethylphosphonate--pyruvate transaminase (TIGR02326; EC 2.6.1.37; HMM-score: 31.5)Biosynthesis of cofactors, prosthetic groups, and carriers Other tyrosine decarboxylase MnfA (TIGR03812; EC 4.1.1.25; HMM-score: 30.8)Amino acid biosynthesis Aspartate family O-acetylhomoserine aminocarboxypropyltransferase/cysteine synthase (TIGR01326; HMM-score: 27.5)Amino acid biosynthesis Serine family O-acetylhomoserine aminocarboxypropyltransferase/cysteine synthase (TIGR01326; HMM-score: 27.5)Amino acid biosynthesis Aspartate family O-succinylhomoserine (thiol)-lyase (TIGR02080; EC 2.5.1.48; HMM-score: 27.2)Protein synthesis tRNA aminoacylation L-seryl-tRNA(Sec) selenium transferase (TIGR00474; EC 2.9.1.1; HMM-score: 25.6)Cellular processes Biosynthesis of natural products capreomycidine synthase (TIGR03947; HMM-score: 25.2)Amino acid biosynthesis Histidine family histidinol-phosphate transaminase (TIGR01141; EC 2.6.1.9; HMM-score: 23)cystathionine beta-lyase (TIGR01329; EC 4.4.1.8; HMM-score: 22.5)putative pyridoxal phosphate-dependent acyltransferase (TIGR01825; EC 2.3.1.-; HMM-score: 22)Cell envelope Biosynthesis and degradation of surface polysaccharides and lipopolysaccharides TDP-4-keto-6-deoxy-D-glucose transaminase (TIGR02379; HMM-score: 21)Amino acid biosynthesis Aspartate family cystathionine beta-lyase (TIGR01324; EC 4.4.1.8; HMM-score: 18)Energy metabolism Amino acids and amines tyrosine aminotransferase (TIGR01264; EC 2.6.1.5; HMM-score: 16.4)UDP-4-amino-4,6-dideoxy-N-acetyl-beta-L-altrosamine transaminase (TIGR03588; EC 2.6.1.92; HMM-score: 16)Energy metabolism Amino acids and amines glycine C-acetyltransferase (TIGR01822; EC 2.3.1.29; HMM-score: 15.2)Biosynthesis of cofactors, prosthetic groups, and carriers Biotin 8-amino-7-oxononanoate synthase (TIGR00858; EC 2.3.1.47; HMM-score: 15.1)Unknown function Enzymes of unknown specificity pyridoxal phosphate enzyme, MJ0158 family (TIGR03576; HMM-score: 15)tyrosine/nicotianamine family aminotransferase (TIGR01265; HMM-score: 14.7)Biosynthesis of cofactors, prosthetic groups, and carriers Heme, porphyrin, and cobalamin 5-aminolevulinic acid synthase (TIGR01821; EC 2.3.1.37; HMM-score: 14)
- TheSEED :
- Cysteine desulfurase (EC 2.8.1.7), SufS subfamily
Amino Acids and Derivatives Alanine, serine, and glycine Alanine biosynthesis Cysteine desulfurase (EC 2.8.1.7), SufS subfamilyand 1 more - PFAM: PLP_aminotran (CL0061) Aminotran_5; Aminotransferase class-V (PF00266; HMM-score: 549.8)and 7 moreCys_Met_Meta_PP; Cys/Met metabolism PLP-dependent enzyme (PF01053; HMM-score: 45.3)DegT_DnrJ_EryC1; DegT/DnrJ/EryC1/StrS aminotransferase family (PF01041; HMM-score: 38.6)Aminotran_1_2; Aminotransferase class I and II (PF00155; HMM-score: 35.3)Beta_elim_lyase; Beta-eliminating lyase (PF01212; HMM-score: 33.5)SelA; L-seryl-tRNA selenium transferase (PF03841; HMM-score: 20.9)Pyridoxal_deC; Pyridoxal-dependent decarboxylase conserved domain (PF00282; HMM-score: 16.7)OKR_DC_1; Orn/Lys/Arg decarboxylase, major domain (PF01276; HMM-score: 11.8)
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors: pyridoxal 5'-phosphate
- effectors:
⊟Localization[edit | edit source]
- PSORTb: Cytoplasmic
- Cytoplasmic Score: 9.97
- Cytoplasmic Membrane Score: 0
- Cellwall Score: 0.01
- Extracellular Score: 0.02
- Internal Helices: 0
- DeepLocPro: Cytoplasmic
- Cytoplasmic Score: 0.997
- Cytoplasmic Membrane Score: 0.0022
- Cell wall & surface Score: 0.0001
- Extracellular Score: 0.0007
- LocateP: Intracellular
- Prediction by SwissProt Classification: Cytoplasmic
- Pathway Prediction: No pathway
- Intracellular possibility: 1
- Signal peptide possibility: -1
- N-terminally Anchored Score: 1
- Predicted Cleavage Site: No CleavageSite
- SignalP: no predicted signal peptide
- SP(Sec/SPI): 0.006062
- TAT(Tat/SPI): 0.000698
- LIPO(Sec/SPII): 0.001239
- predicted transmembrane helices (TMHMM): 0
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MNEVAEHSFDVNEVIKDFPILDQKVNGKRLAYLDSTATSQTPVQVLNVLEDYYKRYNSNVHRGVHTLGSLATDGYENARETVRRFINAKYFEEIIFTRGTTASINLVAHSYGDANVEEGDEIVVTEMEHHANIVPWQQLAKRKNATLKFIPMTADGELNIEDIKQTINDKTKIVAIAHISNVLGTINDVKTIAEIAHQHGAIISVDGAQAAPHMKLDMQEMNADFYSFSGHKMLGPTGIGVLFGKRELLQKMEPIEFGGDMIDFVSKYDATWADLPTKFEAGTPLIAQAIGLAEAIRYLERIGFDAIHKYEQELTIYAYEQMSAIEGIEIYGPPKDRRAGVITFNLQDVHPHDVATAVDTEGVAVRAGHHCAQPLMKWLNVSSTARASFYIYNTKEDVDQLINALKQTKEFFSYEF
⊟Experimental data[edit | edit source]
- experimentally validated: PeptideAtlas [2] [3]
- protein localization: data available for COL
- quantitative data / protein copy number per cell: data available for COL
- interaction partners:
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
- MicrobesOnline: SAOUHSC_00847 > SAOUHSC_00848 > SAOUHSC_00849 > SAOUHSC_00850 > SAOUHSC_00851predicted SigB promoter [4] : SAOUHSC_00845 > S351 > S353 > SAOUHSC_00847 > S354 > SAOUHSC_00848 > S355 > SAOUHSC_00849 > SAOUHSC_00850 > S356 > SAOUHSC_00851 > S357predicted SigB promoter [4] : S353 > SAOUHSC_00847 > S354 > SAOUHSC_00848 > S355 > SAOUHSC_00849 > SAOUHSC_00850 > S356 > SAOUHSC_00851 > S357
⊟Regulation[edit | edit source]
- regulator: PerR* (repression) regulon
PerR* (TF) important in Oxidative stress response; RegPrecise transcription unit transferred from N315 data RegPrecise
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: [4]
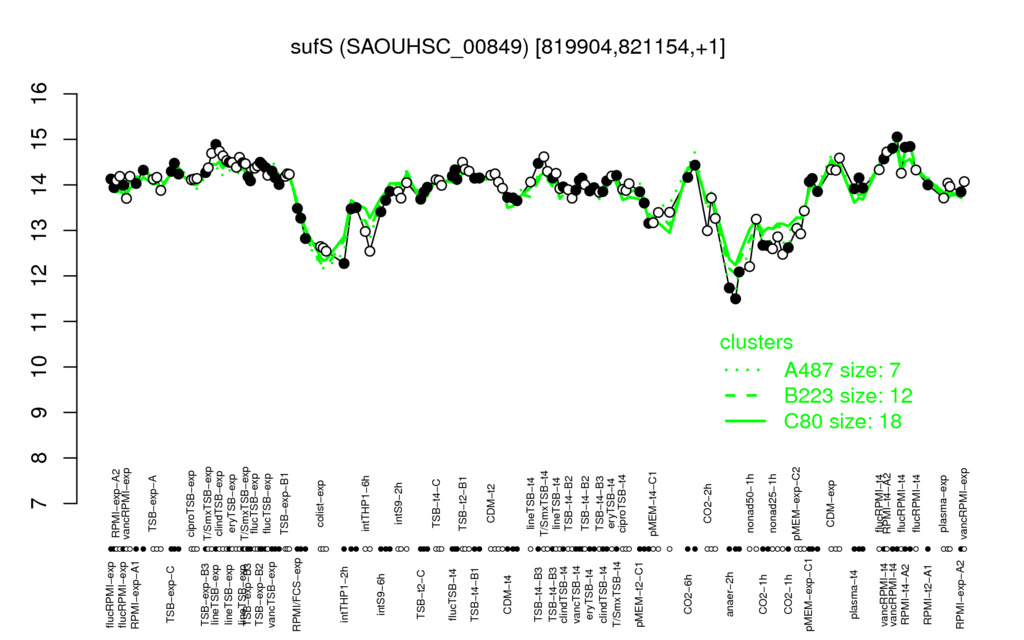 Multi-gene expression profiles
Multi-gene expression profiles
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
⊟Protein stability[edit | edit source]
- half-life: no data available
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You can add further information about the gene and protein here. [edit]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ Roy R Chaudhuri, Andrew G Allen, Paul J Owen, Gil Shalom, Karl Stone, Marcus Harrison, Timothy A Burgis, Michael Lockyer, Jorge Garcia-Lara, Simon J Foster, Stephen J Pleasance, Sarah E Peters, Duncan J Maskell, Ian G Charles
Comprehensive identification of essential Staphylococcus aureus genes using Transposon-Mediated Differential Hybridisation (TMDH).
BMC Genomics: 2009, 10;291
[PubMed:19570206] [WorldCat.org] [DOI] (I e) - ↑ Maren Depke, Stephan Michalik, Alexander Rabe, Kristin Surmann, Lars Brinkmann, Nico Jehmlich, Jörg Bernhardt, Michael Hecker, Bernd Wollscheid, Zhi Sun, Robert L Moritz, Uwe Völker, Frank Schmidt
A peptide resource for the analysis of Staphylococcus aureus in host-pathogen interaction studies.
Proteomics: 2015, 15(21);3648-61
[PubMed:26224020] [WorldCat.org] [DOI] (I p) - ↑ Stephan Michalik, Maren Depke, Annette Murr, Manuela Gesell Salazar, Ulrike Kusebauch, Zhi Sun, Tanja C Meyer, Kristin Surmann, Henrike Pförtner, Petra Hildebrandt, Stefan Weiss, Laura Marcela Palma Medina, Melanie Gutjahr, Elke Hammer, Dörte Becher, Thomas Pribyl, Sven Hammerschmidt, Eric W Deutsch, Samuel L Bader, Michael Hecker, Robert L Moritz, Ulrike Mäder, Uwe Völker, Frank Schmidt
A global Staphylococcus aureus proteome resource applied to the in vivo characterization of host-pathogen interactions.
Sci Rep: 2017, 7(1);9718
[PubMed:28887440] [WorldCat.org] [DOI] (I e) - ↑ 4.0 4.1 4.2 Ulrike Mäder, Pierre Nicolas, Maren Depke, Jan Pané-Farré, Michel Debarbouille, Magdalena M van der Kooi-Pol, Cyprien Guérin, Sandra Dérozier, Aurelia Hiron, Hanne Jarmer, Aurélie Leduc, Stephan Michalik, Ewoud Reilman, Marc Schaffer, Frank Schmidt, Philippe Bessières, Philippe Noirot, Michael Hecker, Tarek Msadek, Uwe Völker, Jan Maarten van Dijl
Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions.
PLoS Genet: 2016, 12(4);e1005962
[PubMed:27035918] [WorldCat.org] [DOI] (I e)
