Jump to navigation
Jump to search
NCBI: 03-AUG-2016
⊟Summary[edit | edit source]
- organism: Staphylococcus aureus NCTC8325
- locus tag: SAOUHSC_02312
- pan locus tag?: SAUPAN005349000
- symbol: SAOUHSC_02312
- pan gene symbol?: kdpA
- synonym:
- product: potassium-transporting ATPase subunit A
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
- type: CDS
- locus tag: SAOUHSC_02312
- symbol: SAOUHSC_02312
- product: potassium-transporting ATPase subunit A
- replicon: chromosome
- strand: -
- coordinates: 2142498..2144174
- length: 1677
- essential: no DEG other strains
⊟Accession numbers[edit | edit source]
- Gene ID: 3920937 NCBI
- RefSeq: YP_500791 NCBI
- BioCyc: G1I0R-2184 BioCyc
- MicrobesOnline: 1290752 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421
481
541
601
661
721
781
841
901
961
1021
1081
1141
1201
1261
1321
1381
1441
1501
1561
1621ATGGAAATAATTTTGTTTCTAACAATGATGGTTATGATTACGTATGTTTTTAGTGGATAT
CTATATCGCGTAGCATTGGTGCAATCGTCAAGAGTAGATTTAATATTTACTAGATTTGAA
AATATGTGTTTTAAAATTATCGGCACAGATTTAGAACACATGTCAGCTAAAACATATGTC
AAACATTTTCTGGCATTTAATGGATTTATGGGGTTTATAACATTTGTATTGCTAATAGTT
CAGCAATGGCTTTTTTTAAATCCAAATCATAATTTAAACCAATCGATAGATTTAGCGTTT
AATACAGCAATATCTTTTTTAACAAATAGTAATTTACAACACTATAACGGTGAATCAGAT
GTGACATATTTAACGCAAATGATTGTAATGACATATTTAATGTTTACATCTAGTGCATCA
GGTTACGCCGTTTGTATAGCGATGTTGAGACGTTTAACTGGATTAACTAATATCATTGGT
AATTTTTATCAAGATATTGTTCGGTTTATTGTCCGAGTACTTTTACCATTATCATGTTTA
ATTAGTATTTTATTGATGACTCAAGGTGTACCACAAACGTTGCATGCTAATTTAATGATT
CGGACTTTAAGCGGACATATTCAACATATTGCATTTGGACCTATTGCATCACTTGAATCA
ATAAAACATCTTGGTACGAATGGTGGAGGATTTTTAGCAGGAAATTCTGCAACACCTTTT
GAAAATCCAAATATTTGGAGCAATTTTATAGAAATGGGCAGTATGATGTTACTTCCTATG
TCAATGTTGTTCTTATTTGGTCGCATGTTAAGTAGACATGGTAAACGAGTACATCGTCAT
GCGTTGATATTATTTGTCGCAATGTTTTTCATTTTTATAGCAATTCTTACATTAACTATG
TGGAGTGAGTATCGTGGTAATCCAATACTAGCGAATTTAGGCATTTATGGACCGAATATG
GAAGGTAAAGAGGTACGGTTTGGAGCAGGTTTGTCAGCACTATTTACAGTTATTACGACG
GCATTTACAACGGGTTCTGTTAATAACATGCATGATAGCTTAACGCCTATAGGTGGATTA
GGACCAATGGTATTAATGATGCTAAATGTTGTATTTGGTGGCGAAGGCGTAGGACTCATG
AATTTATTGATATTTGTCTTACTGACGGTGTTTATATGCAGTTTGATGGTTGGTAAAACA
CCAGAATATTTAAATATGCCAATTGGCGCCCGTGAAATGAAATGTATTGTCTTAGTCTTT
CTCATACACCCAATTTTAATTTTAGTATTTTCAGCACTTGCTTTTATGATTCCTGGAGCA
AGTGAAAGTATAACGAATCCGTCTTTTCATGGTATTTCACAAGTTATGTATGAAATGACA
TCAGCTGCTGCGAACAATGGATCAGGGTTTGAAGGACTGAAAGATGATACAACATTCTGG
AATATCTCTACAGGAATCATTATGTTGCTTTCTCGTTATATACCAATTATTTTGCAATTA
ATGATTGCATCAAGTTTAGTGAATAAAAAATCATACCATCAAGATAAATATACAATAGCG
ATTGATAAACCTTATTTTGGCGTATCACTAATCGTATTTATCGTTTTACTAAGTGGTTTG
ACATTTATTCCAGTACTATTACTTGGTCCTATTGGTGAATTTTTAACTTTAAAATAA60
120
180
240
300
360
420
480
540
600
660
720
780
840
900
960
1020
1080
1140
1200
1260
1320
1380
1440
1500
1560
1620
1677
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SAOUHSC_02312
- symbol: SAOUHSC_02312
- description: potassium-transporting ATPase subunit A
- length: 558
- theoretical pI: 8.44178
- theoretical MW: 62025.5
- GRAVY: 0.688351
⊟Function[edit | edit source]
- reaction: EC 3.6.3.12? ExPASyPotassium-transporting ATPase ATP + H2O + K+(Out) = ADP + phosphate + K+(In)
- TIGRFAM: Transport and binding proteins Cations and iron carrying compounds K+-transporting ATPase, A subunit (TIGR00680; EC 3.6.3.12; HMM-score: 580.7)
- TheSEED :
- Potassium-transporting ATPase A chain (EC 3.6.3.12) (TC 3.A.3.7.1)
- PFAM: Ion_channel (CL0030) KdpA; Potassium-transporting ATPase A subunit (PF03814; HMM-score: 705.4)and 1 moreno clan defined DUF5826; Family of unknown function (DUF5826) (PF19144; HMM-score: 15.3)
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors:
- effectors:
⊟Localization[edit | edit source]
- PSORTb: Cytoplasmic Membrane
- Cytoplasmic Score: 0
- Cytoplasmic Membrane Score: 10
- Cellwall Score: 0
- Extracellular Score: 0
- Internal Helices: 12
- DeepLocPro: Cytoplasmic Membrane
- Cytoplasmic Score: 0
- Cytoplasmic Membrane Score: 0.9999
- Cell wall & surface Score: 0
- Extracellular Score: 0.0001
- LocateP: Multi-transmembrane
- Prediction by SwissProt Classification: Membrane
- Pathway Prediction: Sec-(SPI)
- Intracellular possibility: 0.17
- Signal peptide possibility: -1
- N-terminally Anchored Score: 1
- Predicted Cleavage Site: No CleavageSite
- SignalP: no predicted signal peptide
- SP(Sec/SPI): 0.087662
- TAT(Tat/SPI): 0.001003
- LIPO(Sec/SPII): 0.017459
- predicted transmembrane helices (TMHMM): 12
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MEIILFLTMMVMITYVFSGYLYRVALVQSSRVDLIFTRFENMCFKIIGTDLEHMSAKTYVKHFLAFNGFMGFITFVLLIVQQWLFLNPNHNLNQSIDLAFNTAISFLTNSNLQHYNGESDVTYLTQMIVMTYLMFTSSASGYAVCIAMLRRLTGLTNIIGNFYQDIVRFIVRVLLPLSCLISILLMTQGVPQTLHANLMIRTLSGHIQHIAFGPIASLESIKHLGTNGGGFLAGNSATPFENPNIWSNFIEMGSMMLLPMSMLFLFGRMLSRHGKRVHRHALILFVAMFFIFIAILTLTMWSEYRGNPILANLGIYGPNMEGKEVRFGAGLSALFTVITTAFTTGSVNNMHDSLTPIGGLGPMVLMMLNVVFGGEGVGLMNLLIFVLLTVFICSLMVGKTPEYLNMPIGAREMKCIVLVFLIHPILILVFSALAFMIPGASESITNPSFHGISQVMYEMTSAAANNGSGFEGLKDDTTFWNISTGIIMLLSRYIPIILQLMIASSLVNKKSYHQDKYTIAIDKPYFGVSLIVFIVLLSGLTFIPVLLLGPIGEFLTLK
⊟Experimental data[edit | edit source]
- experimentally validated:
- protein localization:
- quantitative data / protein copy number per cell:
- interaction partners:
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
⊟Regulation[edit | edit source]
- regulator:
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: [1]
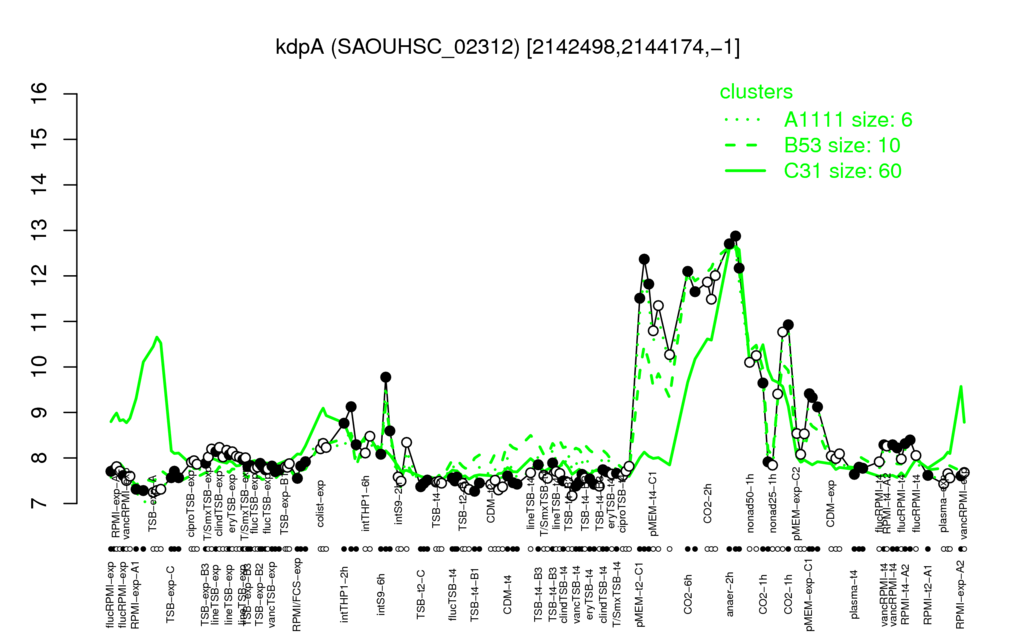 Multi-gene expression profiles
Multi-gene expression profiles
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
- Aureolib: no data available
⊟Protein stability[edit | edit source]
- half-life: no data available
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You can add further information about the gene and protein here. [edit]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ 1.0 1.1 Ulrike Mäder, Pierre Nicolas, Maren Depke, Jan Pané-Farré, Michel Debarbouille, Magdalena M van der Kooi-Pol, Cyprien Guérin, Sandra Dérozier, Aurelia Hiron, Hanne Jarmer, Aurélie Leduc, Stephan Michalik, Ewoud Reilman, Marc Schaffer, Frank Schmidt, Philippe Bessières, Philippe Noirot, Michael Hecker, Tarek Msadek, Uwe Völker, Jan Maarten van Dijl
Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions.
PLoS Genet: 2016, 12(4);e1005962
[PubMed:27035918] [WorldCat.org] [DOI] (I e)
