Jump to navigation
Jump to search
NCBI: 03-AUG-2016
⊟Summary[edit | edit source]
- organism: Staphylococcus aureus NCTC8325
- locus tag: SAOUHSC_02608
- pan locus tag?: SAUPAN005819000
- symbol: SAOUHSC_02608
- pan gene symbol?: hutR
- synonym:
- product: hypothetical protein
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
- type: CDS
- locus tag: SAOUHSC_02608
- symbol: SAOUHSC_02608
- product: hypothetical protein
- replicon: chromosome
- strand: +
- coordinates: 2398225..2399109
- length: 885
- essential: no DEG other strains
⊟Accession numbers[edit | edit source]
- Gene ID: 3921386 NCBI
- RefSeq: YP_501069 NCBI
- BioCyc: G1I0R-2460 BioCyc
- MicrobesOnline: 1291040 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421
481
541
601
661
721
781
841ATGAAAATTATTCAGTTAGAATACTTCTTGGCTATCGTGAAATATAATAGTTTTACTAAA
GCTGCACAATTTTTACATATTAGCCAGCCATCTTTAACTGCTACGATTAAAAAAATGGAA
GCAGATTTAGGTTATGACTTATTTACACGTTCAACAAAAGACATCAAGATTACCGAAAAA
GGAATACAGTTTTATCGTTATGCGAGCGAATTAGTTCAACAATATCGATCCACGATGGAA
AAAATGTATGATTTAAGCGTTACATCAGAACCAAGGATAAAAATTGGGACTCTTGAATCT
ACGAATCAATGGATTGCGAATTTAATTCGAAAGCACCATTCCGACTACCCTGAACAGCAA
TATCGTTTATATGAAATACATGATAAACATCAATCTATAGAGCAATTACTGAATTTTAAT
ATTCATTTAGCTATAACAAATGAAAAAATAACCCACGAAGATATAAGATCCATTCCTTTA
TATGAGGAATCTTACATTTTATTAGCACCCAAGGAAACATTTAAAAATCAAAATTGGGTA
GATGTTGAAAATTTGCCACTCATATTACCAAACAAAAATTCTCAAGTGCGCAAACACTTA
GATGACTATTTTAATAGAAGAAATATTCGTCCAAATGTCGTTGTAGAAACAGATCGATTC
GAATCAGCAGTTGGATTTGTTCATCTCGGCTTAGGTTACGCTATCATTCCGAGATTTTAT
TACCAATCATTTCACACGTCTAATTTAGAATATAAAAAAATTCGTCCAAACTTAGGCCGA
AAAATTTATATCAATTACCATAAAAAACGCAAACACTCCGAACAAGTACATACATTCGTA
CAACAATGCCAAGATTATTTATATGGACTTTTAGAGGCTCTTTAA60
120
180
240
300
360
420
480
540
600
660
720
780
840
885
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SAOUHSC_02608
- symbol: SAOUHSC_02608
- description: hypothetical protein
- length: 294
- theoretical pI: 9.38669
- theoretical MW: 35017.8
- GRAVY: -0.546939
⊟Function[edit | edit source]
- TIGRFAM: Regulatory functions DNA interactions aminoethylphosphonate catabolism associated LysR family transcriptional regulator (TIGR03339; HMM-score: 94.6)Energy metabolism Other pca operon transcription factor PcaQ (TIGR02424; HMM-score: 84.3)Regulatory functions DNA interactions pca operon transcription factor PcaQ (TIGR02424; HMM-score: 84.3)and 6 moreCellular processes Toxin production and resistance transcriptional regulator, ArgP family (TIGR03298; HMM-score: 56.5)DNA metabolism DNA replication, recombination, and repair transcriptional regulator, ArgP family (TIGR03298; HMM-score: 56.5)Regulatory functions DNA interactions transcriptional regulator, ArgP family (TIGR03298; HMM-score: 56.5)putative choline sulfate-utilization transcription factor (TIGR03418; HMM-score: 43.9)Regulatory functions DNA interactions D-serine deaminase transcriptional activator (TIGR02036; HMM-score: 26.9)homoprotocatechuate degradation operon regulator, HpaR (TIGR02337; HMM-score: 13.4)
- TheSEED :
- Transcriptional regulator, LysR family
- PFAM: PBP (CL0177) LysR_substrate; LysR substrate binding domain (PF03466; HMM-score: 123.7)and 4 moreHTH (CL0123) HTH_1; Bacterial regulatory helix-turn-helix protein, lysR family (PF00126; HMM-score: 67.2)NUMOD1; NUMOD1 domain (PF07453; HMM-score: 15.1)MmeI_HD-like (CL0884) HsdM_N; HsdM N-terminal domain (PF12161; HMM-score: 15.1)CheY (CL0304) TadZ-like_ARD; TadZ-like, receiver domain (PF21194; HMM-score: 13.4)
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors:
- effector: Histidine
- genes regulated by HutR*, TF important in Histidine utilizationRegPrecisetranscription units transferred from N315 data RegPrecise
⊟Localization[edit | edit source]
- PSORTb: Cytoplasmic
- Cytoplasmic Score: 9.97
- Cytoplasmic Membrane Score: 0
- Cellwall Score: 0.01
- Extracellular Score: 0.02
- Internal Helices: 0
- DeepLocPro: Cytoplasmic
- Cytoplasmic Score: 0.9196
- Cytoplasmic Membrane Score: 0.0688
- Cell wall & surface Score: 0.0008
- Extracellular Score: 0.0108
- LocateP: Intracellular
- Prediction by SwissProt Classification: Cytoplasmic
- Pathway Prediction: No pathway
- Intracellular possibility: 1
- Signal peptide possibility: -1
- N-terminally Anchored Score: -1
- Predicted Cleavage Site: No CleavageSite
- SignalP: no predicted signal peptide
- SP(Sec/SPI): 0.039007
- TAT(Tat/SPI): 0.004353
- LIPO(Sec/SPII): 0.002704
- predicted transmembrane helices (TMHMM): 0
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MKIIQLEYFLAIVKYNSFTKAAQFLHISQPSLTATIKKMEADLGYDLFTRSTKDIKITEKGIQFYRYASELVQQYRSTMEKMYDLSVTSEPRIKIGTLESTNQWIANLIRKHHSDYPEQQYRLYEIHDKHQSIEQLLNFNIHLAITNEKITHEDIRSIPLYEESYILLAPKETFKNQNWVDVENLPLILPNKNSQVRKHLDDYFNRRNIRPNVVVETDRFESAVGFVHLGLGYAIIPRFYYQSFHTSNLEYKKIRPNLGRKIYINYHKKRKHSEQVHTFVQQCQDYLYGLLEAL
⊟Experimental data[edit | edit source]
- experimentally validated: PeptideAtlas [1] [2]
- protein localization: data available for COL
- quantitative data / protein copy number per cell:
- interaction partners:
SAOUHSC_01170 (carB) carbamoyl phosphate synthase large subunit [3] (data from MRSA252) SAOUHSC_01683 (dnaK) molecular chaperone DnaK [3] (data from MRSA252) SAOUHSC_00799 (eno) phosphopyruvate hydratase [3] (data from MRSA252) SAOUHSC_02117 (gatA) aspartyl/glutamyl-tRNA amidotransferase subunit A [3] (data from MRSA252) SAOUHSC_02254 (groEL) chaperonin GroEL [3] (data from MRSA252) SAOUHSC_00519 (rplA) 50S ribosomal protein L1 [3] (data from MRSA252) SAOUHSC_02500 (rplE) 50S ribosomal protein L5 [3] (data from MRSA252) SAOUHSC_00518 (rplK) 50S ribosomal protein L11 [3] (data from MRSA252) SAOUHSC_00521 (rplL) 50S ribosomal protein L7/L12 [3] (data from MRSA252) SAOUHSC_02505 (rplP) 50S ribosomal protein L16 [3] (data from MRSA252) SAOUHSC_01211 (rplS) 50S ribosomal protein L19 [3] (data from MRSA252) SAOUHSC_01757 (rplU) 50S ribosomal protein L21 [3] (data from MRSA252) SAOUHSC_02510 (rplW) 50S ribosomal protein L23 [3] (data from MRSA252) SAOUHSC_01232 (rpsB) 30S ribosomal protein S2 [3] (data from MRSA252) SAOUHSC_02494 (rpsE) 30S ribosomal protein S5 [3] (data from MRSA252) SAOUHSC_02498 (rpsH) 30S ribosomal protein S8 [3] (data from MRSA252) SAOUHSC_00769 (secA) preprotein translocase subunit SecA [3] (data from MRSA252) SAOUHSC_01234 (tsf) elongation factor Ts [3] (data from MRSA252) SAOUHSC_02353 (upp) uracil phosphoribosyltransferase [3] (data from MRSA252) SAOUHSC_00187 formate acetyltransferase [3] (data from MRSA252) SAOUHSC_00206 L-lactate dehydrogenase [3] (data from MRSA252) SAOUHSC_00336 acetyl-CoA acyltransferase [3] (data from MRSA252) SAOUHSC_00472 ribose-phosphate pyrophosphokinase [3] (data from MRSA252) SAOUHSC_00528 30S ribosomal protein S7 [3] (data from MRSA252) SAOUHSC_00529 elongation factor G [3] (data from MRSA252) SAOUHSC_00530 elongation factor Tu [3] (data from MRSA252) SAOUHSC_00535 hypothetical protein [3] (data from MRSA252) SAOUHSC_00679 hypothetical protein [3] (data from MRSA252) SAOUHSC_00690 hypothetical protein [3] (data from MRSA252) SAOUHSC_00742 ribonucleotide-diphosphate reductase subunit alpha [3] (data from MRSA252) SAOUHSC_00795 glyceraldehyde-3-phosphate dehydrogenase [3] (data from MRSA252) SAOUHSC_00947 enoyl-(acyl carrier protein) reductase [3] (data from MRSA252) SAOUHSC_01040 pyruvate dehydrogenase complex, E1 component subunit alpha [3] (data from MRSA252) SAOUHSC_01150 cell division protein FtsZ [3] (data from MRSA252) SAOUHSC_01416 dihydrolipoamide succinyltransferase [3] (data from MRSA252) SAOUHSC_01451 threonine dehydratase [3] (data from MRSA252) SAOUHSC_01490 DNA-binding protein HU [3] (data from MRSA252) SAOUHSC_01605 6-phosphogluconate dehydrogenase [3] (data from MRSA252) SAOUHSC_01801 isocitrate dehydrogenase [3] (data from MRSA252) SAOUHSC_01806 pyruvate kinase [3] (data from MRSA252) SAOUHSC_01819 hypothetical protein [3] (data from MRSA252) SAOUHSC_01820 acetate kinase [3] (data from MRSA252) SAOUHSC_02337 UDP-N-acetylglucosamine 1-carboxyvinyltransferase [3] (data from MRSA252) SAOUHSC_02399 glucosamine--fructose-6-phosphate aminotransferase [3] (data from MRSA252) SAOUHSC_02441 alkaline shock protein 23 [3] (data from MRSA252) SAOUHSC_02926 fructose-1,6-bisphosphate aldolase [3] (data from MRSA252) SAOUHSC_02969 arginine deiminase [3] (data from MRSA252)
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
- MicrobesOnline: no polycistronic organisation predicted
⊟Regulation[edit | edit source]
- regulator: HutR* (activation) regulon
HutR* (TF) important in Histidine utilization; RegPrecise
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: [4]
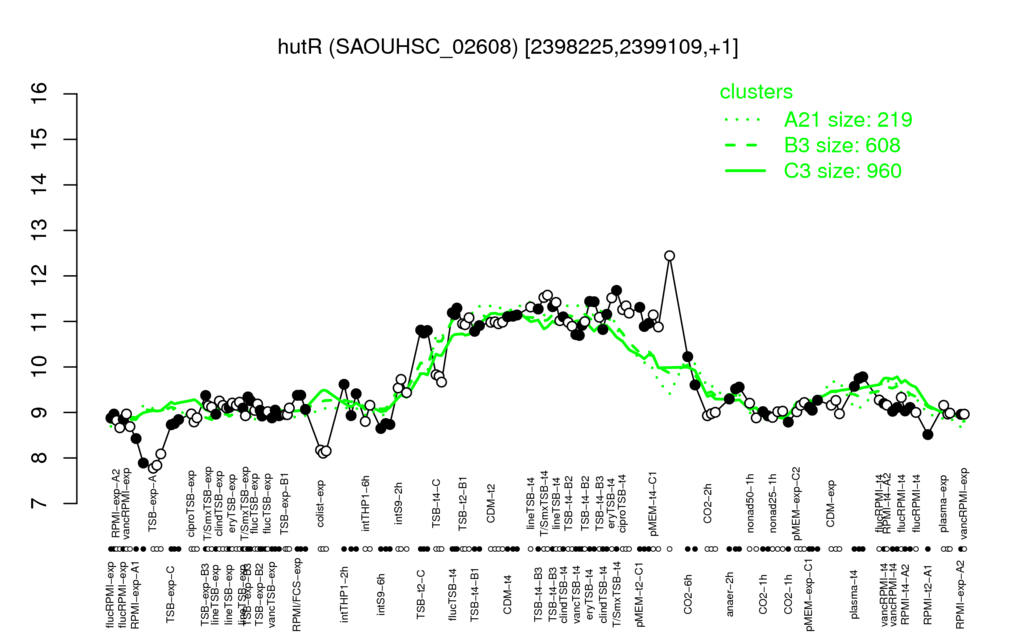 Multi-gene expression profiles
Multi-gene expression profiles
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
- Aureolib: no data available
⊟Protein stability[edit | edit source]
- half-life: no data available
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You can add further information about the gene and protein here. [edit]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ Maren Depke, Stephan Michalik, Alexander Rabe, Kristin Surmann, Lars Brinkmann, Nico Jehmlich, Jörg Bernhardt, Michael Hecker, Bernd Wollscheid, Zhi Sun, Robert L Moritz, Uwe Völker, Frank Schmidt
A peptide resource for the analysis of Staphylococcus aureus in host-pathogen interaction studies.
Proteomics: 2015, 15(21);3648-61
[PubMed:26224020] [WorldCat.org] [DOI] (I p) - ↑ Stephan Michalik, Maren Depke, Annette Murr, Manuela Gesell Salazar, Ulrike Kusebauch, Zhi Sun, Tanja C Meyer, Kristin Surmann, Henrike Pförtner, Petra Hildebrandt, Stefan Weiss, Laura Marcela Palma Medina, Melanie Gutjahr, Elke Hammer, Dörte Becher, Thomas Pribyl, Sven Hammerschmidt, Eric W Deutsch, Samuel L Bader, Michael Hecker, Robert L Moritz, Ulrike Mäder, Uwe Völker, Frank Schmidt
A global Staphylococcus aureus proteome resource applied to the in vivo characterization of host-pathogen interactions.
Sci Rep: 2017, 7(1);9718
[PubMed:28887440] [WorldCat.org] [DOI] (I e) - ↑ 3.00 3.01 3.02 3.03 3.04 3.05 3.06 3.07 3.08 3.09 3.10 3.11 3.12 3.13 3.14 3.15 3.16 3.17 3.18 3.19 3.20 3.21 3.22 3.23 3.24 3.25 3.26 3.27 3.28 3.29 3.30 3.31 3.32 3.33 3.34 3.35 3.36 3.37 3.38 3.39 3.40 3.41 3.42 3.43 3.44 3.45 3.46 Artem Cherkasov, Michael Hsing, Roya Zoraghi, Leonard J Foster, Raymond H See, Nikolay Stoynov, Jihong Jiang, Sukhbir Kaur, Tian Lian, Linda Jackson, Huansheng Gong, Rick Swayze, Emily Amandoron, Farhad Hormozdiari, Phuong Dao, Cenk Sahinalp, Osvaldo Santos-Filho, Peter Axerio-Cilies, Kendall Byler, William R McMaster, Robert C Brunham, B Brett Finlay, Neil E Reiner
Mapping the protein interaction network in methicillin-resistant Staphylococcus aureus.
J Proteome Res: 2011, 10(3);1139-50
[PubMed:21166474] [WorldCat.org] [DOI] (I p) - ↑ Ulrike Mäder, Pierre Nicolas, Maren Depke, Jan Pané-Farré, Michel Debarbouille, Magdalena M van der Kooi-Pol, Cyprien Guérin, Sandra Dérozier, Aurelia Hiron, Hanne Jarmer, Aurélie Leduc, Stephan Michalik, Ewoud Reilman, Marc Schaffer, Frank Schmidt, Philippe Bessières, Philippe Noirot, Michael Hecker, Tarek Msadek, Uwe Völker, Jan Maarten van Dijl
Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions.
PLoS Genet: 2016, 12(4);e1005962
[PubMed:27035918] [WorldCat.org] [DOI] (I e)
