Jump to navigation
Jump to search
NCBI: 10-JUN-2013
⊟Summary[edit | edit source]
- organism: Staphylococcus aureus COL
- locus tag: SACOL1685 [new locus tag: SACOL_RS08595 ]
- pan locus tag?: SAUPAN004235000
- symbol: aspS
- pan gene symbol?: aspS
- synonym:
- product: aspartyl-tRNA synthetase
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
- type: CDS
- locus tag: SACOL1685 [new locus tag: SACOL_RS08595 ]
- symbol: aspS
- product: aspartyl-tRNA synthetase
- replicon: chromosome
- strand: -
- coordinates: 1713268..1715034
- length: 1767
- essential: unknown other strains
⊟Accession numbers[edit | edit source]
- Gene ID: 3238249 NCBI
- RefSeq: YP_186524 NCBI
- BioCyc: see SACOL_RS08595
- MicrobesOnline: 913133 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421
481
541
601
661
721
781
841
901
961
1021
1081
1141
1201
1261
1321
1381
1441
1501
1561
1621
1681
1741ATGAGTAAGAGAACAACTTATTGTGGATTAGTTACTGAGGCATTTTTAGGACAAGAAATT
ACATTAAAAGGATGGGTTAACAATCGTCGTGACCTTGGTGGATTGATTTTCGTTGATTTA
AGAGATAGAGAAGGAATTGTACAAGTCGTGTTTAATCCTGCATTTTCAGAAGAGGCATTG
AAAATTGCTGAAACAGTACGTTCTGAATATGTTGTAGAAGTTCAAGGTACAGTTACGAAG
CGTGACCCTGAAACAGTTAATCCTAAAATTAAAACTGGCCAAGTTGAAGTACAAGTTACA
AATATTAAAGTGATTAATAAATCTGAGACACCACCATTTTCTATAAATGAAGAAAATGTT
AACGTTGATGAAAATATTCGATTAAAATACCGTTATTTAGATTTACGTCGTCAAGAGTTA
GCGCAAACATTTAAAATGAGACATCAAATTACACGTTCTATTCGTCAATATTTGGATGAT
GAAGGGTTCTTTGACATCGAAACACCAGTACTAACGAAGTCAACACCTGAGGGTGCACGT
GACTATTTAGTACCATCTCGTGTTCATGATGGTGAATTTTATGCATTACCACAATCACCA
CAATTATTTAAGCAATTATTGATGATTAGTGGATTTGACAAATACTACCAAATCGTAAAA
TGCTTCCGTGACGAAGATTTACGTGCAGATCGTCAACCTGAATTTACACAAGTCGATATT
GAAATGAGTTTTGTAGACCAAGAAGATGTGATGCAAATGGGTGAAGAAATGCTTAAAAAA
GTTGTTAAAGAAGTTAAAGGCGTTGAAATTAATGGCGCTTTCCCACGCATGACATATAAA
GAAGCGATGCGTCGCTATGGTTCTGATAAACCAGATACACGTTTTGAAATGGAATTAATT
GACGTTTCTCAATTAGGACGTGATATGGACTTTAAAGTATTTAAAGATACTGTTGAAAAT
GATGGTGAAATTAAAGCAATTGTCGCTAAAGGTGCAGCTGAACAATATACTCGTAAAGAT
ATGGATGCTTTAACAGAATTTGTAAACATCTATGGTGCTAAAGGATTAGCGTGGGTTAAA
GTTGTGGAAGATGGTTTGACAGGTCCAATTGGACGTTTCTTTGAAACAGAAAATGTTGAA
ACATTACTTACATTAACTGGTGCTGAAGCTGGTGACTTAGTAATGTTTGTTGCAGACAAA
CCAAATGTCGTTGCACAAAGTTTAGGTGCATTACGTGTCAAATTAGCTAAAGAATTAGGT
TTAATCGATGAAACAAAATTAAACTTCTTATGGGTGACAGATTGGCCATTATTAGAATAT
GATGAAGATGCGAAACGTTACGTTGCAGCACATCATCCATTTACATCTCCAAAAGAAGCT
GATATTGCTAAGCTTGGCACTGCGCCAGAAGAAGCTGAGGCAAATGCTTATGACATAGTA
TTAAATGGTTATGAATTAGGTGGCGGTTCAATCAGAATTCATGATGGTGAGTTACAAGAA
AAAATGTTCGAAGTTCTTGGATTTACTAAAGAACAAGCACAAGAACAGTTCGGCTTCTTA
CTAGATGCATTTAAATATGGTGCACCACCACATGGCGGTATTGCATTAGGATTAGACCGA
TTAGTCATGTTATTGACTAATAGAACAAACTTGAGAGATACAATCGCATTCCCTAAAACA
GCATCTGCTACATGTTTATTAACGAATGCGCCTGGTGAAGTTTCTGATAAACAATTAGAA
GAACTTTCTTTGCGAATTCGTCACTAA60
120
180
240
300
360
420
480
540
600
660
720
780
840
900
960
1020
1080
1140
1200
1260
1320
1380
1440
1500
1560
1620
1680
1740
1767
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SACOL1685 [new locus tag: SACOL_RS08595 ]
- symbol: AspS
- description: aspartyl-tRNA synthetase
- length: 588
- theoretical pI: 4.69429
- theoretical MW: 66598.4
- GRAVY: -0.355272
⊟Function[edit | edit source]
- reaction: EC 6.1.1.12? ExPASyAspartate--tRNA ligase ATP + L-aspartate + tRNA(Asp) = AMP + diphosphate + L-aspartyl-tRNA(Asp)
- TIGRFAM: Protein synthesis tRNA aminoacylation aspartate--tRNA ligase (TIGR00459; EC 6.1.1.-; HMM-score: 855.8)and 7 moreProtein synthesis tRNA aminoacylation aspartate--tRNA(Asn) ligase (TIGR00458; EC 6.1.1.23; HMM-score: 253.8)Protein synthesis tRNA aminoacylation asparagine--tRNA ligase (TIGR00457; EC 6.1.1.22; HMM-score: 167.8)Protein synthesis tRNA aminoacylation lysine--tRNA ligase (TIGR00499; EC 6.1.1.6; HMM-score: 166.9)Unknown function General EF-P lysine aminoacylase GenX (TIGR00462; HMM-score: 91.8)Protein synthesis tRNA aminoacylation histidine--tRNA ligase (TIGR00442; EC 6.1.1.21; HMM-score: 24.4)Protein synthesis tRNA aminoacylation pyrrolysine--tRNA ligase, C-terminal region (TIGR02367; EC 6.1.1.26; HMM-score: 16.2)Protein synthesis tRNA aminoacylation phenylalanine--tRNA ligase, alpha subunit (TIGR00468; EC 6.1.1.20; HMM-score: 9.9)
- TheSEED :
- Aspartyl-tRNA synthetase (EC 6.1.1.12)
- PFAM: tRNA_synt_II (CL0040) tRNA-synt_2; tRNA synthetases class II (D, K and N) (PF00152; HMM-score: 444)and 6 moreGAD (CL0250) GAD; GAD domain (PF02938; HMM-score: 98.5)OB (CL0021) tRNA_anti-codon; OB-fold nucleic acid binding domain (PF01336; HMM-score: 69.4)tRNA_synt_II (CL0040) tRNA-synt_2d; tRNA synthetases class II core domain (F) (PF01409; HMM-score: 20.3)LRR (CL0022) LRR_NXF1-5; NXF1/2/3/5 leucine-rich repeat domain (PF24048; HMM-score: 17)tRNA_synt_II (CL0040) tRNA-synt_2b; tRNA synthetase class II core domain (G, H, P, S and T) (PF00587; HMM-score: 14.1)no clan defined INTS7_C; Integrator complex subunit 7, C-terminal domain (PF22965; HMM-score: 11.9)
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors:
- effectors:
⊟Localization[edit | edit source]
- PSORTb: Cytoplasmic
- Cytoplasmic Score: 10
- Cytoplasmic Membrane Score: 0
- Cellwall Score: 0
- Extracellular Score: 0
- Internal Helices: 0
- DeepLocPro: Cytoplasmic
- Cytoplasmic Score: 0.9996
- Cytoplasmic Membrane Score: 0
- Cell wall & surface Score: 0
- Extracellular Score: 0.0004
- LocateP: Intracellular
- Prediction by SwissProt Classification: Cytoplasmic
- Pathway Prediction: No pathway
- Intracellular possibility: 1
- Signal peptide possibility: -1
- N-terminally Anchored Score: 1
- Predicted Cleavage Site: No CleavageSite
- SignalP: no predicted signal peptide
- SP(Sec/SPI): 0.004914
- TAT(Tat/SPI): 0.001285
- LIPO(Sec/SPII): 0.00119
- predicted transmembrane helices (TMHMM): 0
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MSKRTTYCGLVTEAFLGQEITLKGWVNNRRDLGGLIFVDLRDREGIVQVVFNPAFSEEALKIAETVRSEYVVEVQGTVTKRDPETVNPKIKTGQVEVQVTNIKVINKSETPPFSINEENVNVDENIRLKYRYLDLRRQELAQTFKMRHQITRSIRQYLDDEGFFDIETPVLTKSTPEGARDYLVPSRVHDGEFYALPQSPQLFKQLLMISGFDKYYQIVKCFRDEDLRADRQPEFTQVDIEMSFVDQEDVMQMGEEMLKKVVKEVKGVEINGAFPRMTYKEAMRRYGSDKPDTRFEMELIDVSQLGRDMDFKVFKDTVENDGEIKAIVAKGAAEQYTRKDMDALTEFVNIYGAKGLAWVKVVEDGLTGPIGRFFETENVETLLTLTGAEAGDLVMFVADKPNVVAQSLGALRVKLAKELGLIDETKLNFLWVTDWPLLEYDEDAKRYVAAHHPFTSPKEADIAKLGTAPEEAEANAYDIVLNGYELGGGSIRIHDGELQEKMFEVLGFTKEQAQEQFGFLLDAFKYGAPPHGGIALGLDRLVMLLTNRTNLRDTIAFPKTASATCLLTNAPGEVSDKQLEELSLRIRH
⊟Experimental data[edit | edit source]
- experimentally validated: PeptideAtlas
- protein localization: Cytoplasmic [1] [2] [3]
- quantitative data / protein copy number per cell: 1172 [4]
- interaction partners:
SACOL0593 (fusA) elongation factor G [5] (data from MRSA252) SACOL2145 (glmS) glucosamine--fructose-6-phosphate aminotransferase [5] (data from MRSA252) SACOL1741 (icd) isocitrate dehydrogenase [5] (data from MRSA252) SACOL1103 (pdhB) pyruvate dehydrogenase complex E1 component subunit beta [5] (data from MRSA252) SACOL1105 (pdhD) dihydrolipoamide dehydrogenase [5] (data from MRSA252) SACOL1745 (pyk) pyruvate kinase [5] (data from MRSA252) SACOL0589 (rpoC) DNA-directed RNA polymerase subunit beta' [5] (data from MRSA252) SACOL1274 (rpsB) 30S ribosomal protein S2 [5] (data from MRSA252) SACOL2233 (rpsC) 30S ribosomal protein S3 [5] (data from MRSA252) SACOL2222 (rpsE) 30S ribosomal protein S5 [5] (data from MRSA252) SACOL2206 (rpsI) 30S ribosomal protein S9 [5] (data from MRSA252) SACOL1263 (sucD) succinyl-CoA synthetase subunit alpha [5] (data from MRSA252) SACOL0594 (tuf) elongation factor Tu [5] (data from MRSA252) SACOL1759 universal stress protein [5] (data from MRSA252) SACOL2173 alkaline shock protein 23 [5] (data from MRSA252)
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
⊟Regulation[edit | edit source]
- regulator: T-box(Asn) (transcription antitermination) regulon
T-box(Asn) (RNA) important in Amino acid metabolism; transcription unit transferred from N315 data RegPrecise
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: data available for NCTC8325
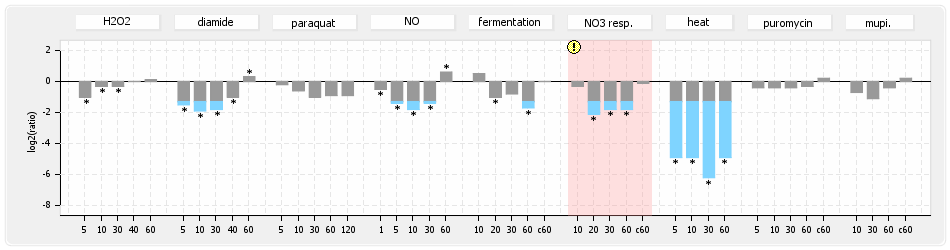
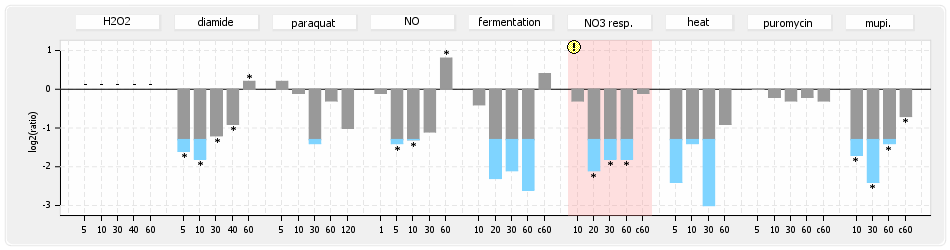
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
⊟Protein stability[edit | edit source]
- half-life: 286.77 h [6]
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You can add further information about the gene and protein here. [edit]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ Dörte Becher, Kristina Hempel, Susanne Sievers, Daniela Zühlke, Jan Pané-Farré, Andreas Otto, Stephan Fuchs, Dirk Albrecht, Jörg Bernhardt, Susanne Engelmann, Uwe Völker, Jan Maarten van Dijl, Michael Hecker
A proteomic view of an important human pathogen--towards the quantification of the entire Staphylococcus aureus proteome.
PLoS One: 2009, 4(12);e8176
[PubMed:19997597] [WorldCat.org] [DOI] (I e) - ↑ Kristina Hempel, Florian-Alexander Herbst, Martin Moche, Michael Hecker, Dörte Becher
Quantitative proteomic view on secreted, cell surface-associated, and cytoplasmic proteins of the methicillin-resistant human pathogen Staphylococcus aureus under iron-limited conditions.
J Proteome Res: 2011, 10(4);1657-66
[PubMed:21323324] [WorldCat.org] [DOI] (I p) - ↑ Andreas Otto, Jan Maarten van Dijl, Michael Hecker, Dörte Becher
The Staphylococcus aureus proteome.
Int J Med Microbiol: 2014, 304(2);110-20
[PubMed:24439828] [WorldCat.org] [DOI] (I p) - ↑ Daniela Zühlke, Kirsten Dörries, Jörg Bernhardt, Sandra Maaß, Jan Muntel, Volkmar Liebscher, Jan Pané-Farré, Katharina Riedel, Michael Lalk, Uwe Völker, Susanne Engelmann, Dörte Becher, Stephan Fuchs, Michael Hecker
Costs of life - Dynamics of the protein inventory of Staphylococcus aureus during anaerobiosis.
Sci Rep: 2016, 6;28172
[PubMed:27344979] [WorldCat.org] [DOI] (I e) - ↑ 5.00 5.01 5.02 5.03 5.04 5.05 5.06 5.07 5.08 5.09 5.10 5.11 5.12 5.13 5.14 Artem Cherkasov, Michael Hsing, Roya Zoraghi, Leonard J Foster, Raymond H See, Nikolay Stoynov, Jihong Jiang, Sukhbir Kaur, Tian Lian, Linda Jackson, Huansheng Gong, Rick Swayze, Emily Amandoron, Farhad Hormozdiari, Phuong Dao, Cenk Sahinalp, Osvaldo Santos-Filho, Peter Axerio-Cilies, Kendall Byler, William R McMaster, Robert C Brunham, B Brett Finlay, Neil E Reiner
Mapping the protein interaction network in methicillin-resistant Staphylococcus aureus.
J Proteome Res: 2011, 10(3);1139-50
[PubMed:21166474] [WorldCat.org] [DOI] (I p) - ↑ Stephan Michalik, Jörg Bernhardt, Andreas Otto, Martin Moche, Dörte Becher, Hanna Meyer, Michael Lalk, Claudia Schurmann, Rabea Schlüter, Holger Kock, Ulf Gerth, Michael Hecker
Life and death of proteins: a case study of glucose-starved Staphylococcus aureus.
Mol Cell Proteomics: 2012, 11(9);558-70
[PubMed:22556279] [WorldCat.org] [DOI] (I p)