Jump to navigation
Jump to search
NCBI: 03-AUG-2016
⊟Summary[edit | edit source]
- organism: Staphylococcus aureus NCTC8325
- locus tag: SAOUHSC_01643
- pan locus tag?: SAUPAN004123000
- symbol: SAOUHSC_01643
- pan gene symbol?: comGA
- synonym:
- product: hypothetical protein
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
- type: CDS
- locus tag: SAOUHSC_01643
- symbol: SAOUHSC_01643
- product: hypothetical protein
- replicon: chromosome
- strand: -
- coordinates: 1559316..1560290
- length: 975
- essential: no DEG other strains
⊟Accession numbers[edit | edit source]
- Gene ID: 3920095 NCBI
- RefSeq: YP_500155 NCBI
- BioCyc: G1I0R-1528 BioCyc
- MicrobesOnline: 1290069 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421
481
541
601
661
721
781
841
901
961TTGAAGATTCTATTTCAAGAAATAATTAATAAAGCGATAGAAATGAAAGCGAGTGATGTA
CATTTTATTCCAGTTAAAAATGAAGTAAGTATTAAATTTAGAATTAATGATAACTTGGAG
CAGTATGAACAAATTGGGAATAGCATTTATCAAAAGTTATTAGTTTATATGAAGTTTCAA
GCTGGGCTTGATGTTTCTACACAGCAAGTCGCACAGAGCGGTCGATATAGTTACCTTTTC
AATAAAATATATTTTTTGAGAATATCAACTTTACCATTGTCACTTGGCCAAGAAAGTTGT
GTTATCAGAATTGTACCTCAATTTTTTCAACCACAGAAATCAACTTATAAATTCAATGAT
TTTAAACACCTCATGAATAAGAAACAAGGATTACTATTGTTTAGTGGACCAACTGGTTCA
GGAAAGAGTACATTAATGTATCAAATGGTCTCATACGCGAATAAAGCCTTGAATTTAAAT
GTAATTTCTATAGAGGATCCTGTAGAGATGCAAATTCCTGGTATCGTCCAAATTAATGTG
AATGATAAAGCTGGCATAAACTATGTAAATTCGTTTAAAGCTATTTTAAGATGTGATCCT
GATGTTATTTTAATAGGTGAAATCAGAGATAAAGATGTTGCCAAGTGTGTTATACAGGCT
AGTTTAAGTGGTCACCTTGTTCTGACTACATTGCATGCAACTGATTGTAAAGGTGCTATT
TTAAGGCTATTAGAAATGGGCATTTCTGTACAAGAATTGATACAGGCAACTAACTTAATT
ATAAACCAACGACTTGTAACTACTATTAAGCAACAGCGACAATTAGTATGTGAAATTCTA
TCTCAGCAACAACTCCGATATTTCTTTTCCCATAATCATTCATTACCATCATCATTTAAG
AACTTAGAAGATAAACTTGATGATATGACAAAAGCAGGTGTCATTTGTGAAACTACAATG
CATAAATACATTTAA60
120
180
240
300
360
420
480
540
600
660
720
780
840
900
960
975
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SAOUHSC_01643
- symbol: SAOUHSC_01643
- description: hypothetical protein
- length: 324
- theoretical pI: 9.07987
- theoretical MW: 36864.8
- GRAVY: -0.0317901
⊟Function[edit | edit source]
- TIGRFAM: Cellular processes Pathogenesis type II secretion system protein E (TIGR02533; HMM-score: 221.8)Protein fate Protein and peptide secretion and trafficking type II secretion system protein E (TIGR02533; HMM-score: 221.8)Cell envelope Surface structures type IV-A pilus assembly ATPase PilB (TIGR02538; HMM-score: 213.5)Protein fate Protein and peptide secretion and trafficking type IV-A pilus assembly ATPase PilB (TIGR02538; HMM-score: 213.5)and 14 moreCell envelope Surface structures twitching motility protein (TIGR01420; HMM-score: 146.5)Cellular processes Chemotaxis and motility twitching motility protein (TIGR01420; HMM-score: 146.5)Mobile and extrachromosomal element functions Plasmid functions plasmid transfer ATPase TraJ (TIGR02525; HMM-score: 84.9)Dot/Icm secretion system ATPase DotB (TIGR02524; HMM-score: 58.2)Cellular processes Conjugation P-type conjugative transfer ATPase TrbB (TIGR02782; HMM-score: 57.5)P-type DNA transfer ATPase VirB11 (TIGR02788; HMM-score: 54.5)helicase/secretion neighborhood ATPase (TIGR03819; HMM-score: 35.9)Cellular processes Sporulation and germination stage III sporulation protein AA (TIGR02858; HMM-score: 17.1)phosphonate C-P lyase system protein PhnL (TIGR02324; HMM-score: 14.7)DNA metabolism DNA replication, recombination, and repair DnaA regulatory inactivator Hda (TIGR03420; HMM-score: 14.4)Protein fate Protein and peptide secretion and trafficking putative secretion ATPase, PEP-CTERM locus subfamily (TIGR03015; HMM-score: 14.1)Cellular processes Chemotaxis and motility flagellar biosynthesis protein FlhF (TIGR03499; HMM-score: 12.9)Purines, pyrimidines, nucleosides, and nucleotides Nucleotide and nucleoside interconversions guanylate kinase (TIGR03263; EC 2.7.4.8; HMM-score: 12.8)Protein synthesis tRNA and rRNA base modification tRNA 2-selenouridine synthase (TIGR03167; EC 2.9.1.-; HMM-score: 12.2)
- TheSEED :
- Late competence protein ComGA, access of DNA to ComEA
DNA Metabolism DNA uptake, competence Gram Positive Competence Late competence protein ComGA, access of DNA to ComEAand 1 more - PFAM: P-loop_NTPase (CL0023) T2SSE; Type II/IV secretion system protein (PF00437; HMM-score: 251.4)and 21 moreAAA_29; P-loop containing region of AAA domain (PF13555; HMM-score: 25.7)nSTAND3; Novel STAND NTPase 3 (PF20720; HMM-score: 19.7)Rad17; Rad17 P-loop domain (PF03215; HMM-score: 18.7)DAP3; Mitochondrial ribosomal death-associated protein 3 (PF10236; HMM-score: 18.2)AAA_7; P-loop containing dynein motor region (PF12775; HMM-score: 18)AAA_14; AAA domain (PF13173; HMM-score: 17.6)NPHP3_N; Nephrocystin 3, N-terminal (PF24883; HMM-score: 17.2)ResIII; Type III restriction enzyme, res subunit (PF04851; HMM-score: 16.3)AAA; ATPase family associated with various cellular activities (AAA) (PF00004; HMM-score: 16.2)AAA_23; AAA domain (PF13476; HMM-score: 15.9)AAA_22; AAA domain (PF13401; HMM-score: 15.2)ATP_bind_1; Conserved hypothetical ATP binding protein (PF03029; HMM-score: 15.1)AAA_30; AAA domain (PF13604; HMM-score: 14.9)RNA_helicase; RNA helicase (PF00910; HMM-score: 14.4)RsgA_GTPase; RsgA GTPase (PF03193; HMM-score: 14.4)G-alpha; G-protein alpha subunit (PF00503; HMM-score: 14)NB-ARC; NB-ARC domain (PF00931; HMM-score: 13.2)cobW; CobW/HypB/UreG, nucleotide-binding domain (PF02492; HMM-score: 13.2)AAA_16; AAA ATPase domain (PF13191; HMM-score: 13)MMR_HSR1; 50S ribosome-binding GTPase (PF01926; HMM-score: 12.5)AAA_25; AAA domain (PF13481; HMM-score: 11.4)
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors:
- effectors:
⊟Localization[edit | edit source]
- PSORTb: Cytoplasmic
- Cytoplasmic Score: 9.89
- Cytoplasmic Membrane Score: 0.09
- Cellwall Score: 0.01
- Extracellular Score: 0.02
- Internal Helices: 0
- DeepLocPro: Cytoplasmic
- Cytoplasmic Score: 0.6324
- Cytoplasmic Membrane Score: 0.3537
- Cell wall & surface Score: 0
- Extracellular Score: 0.0139
- LocateP: Intracellular
- Prediction by SwissProt Classification: Cytoplasmic
- Pathway Prediction: No pathway
- Intracellular possibility: 1
- Signal peptide possibility: -1
- N-terminally Anchored Score: 1
- Predicted Cleavage Site: No CleavageSite
- SignalP: no predicted signal peptide
- SP(Sec/SPI): 0.003912
- TAT(Tat/SPI): 0.000073
- LIPO(Sec/SPII): 0.000327
- predicted transmembrane helices (TMHMM): 0
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MKILFQEIINKAIEMKASDVHFIPVKNEVSIKFRINDNLEQYEQIGNSIYQKLLVYMKFQAGLDVSTQQVAQSGRYSYLFNKIYFLRISTLPLSLGQESCVIRIVPQFFQPQKSTYKFNDFKHLMNKKQGLLLFSGPTGSGKSTLMYQMVSYANKALNLNVISIEDPVEMQIPGIVQINVNDKAGINYVNSFKAILRCDPDVILIGEIRDKDVAKCVIQASLSGHLVLTTLHATDCKGAILRLLEMGISVQELIQATNLIINQRLVTTIKQQRQLVCEILSQQQLRYFFSHNHSLPSSFKNLEDKLDDMTKAGVICETTMHKYI
⊟Experimental data[edit | edit source]
- experimentally validated:
- protein localization:
- quantitative data / protein copy number per cell:
- interaction partners:
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
⊟Regulation[edit | edit source]
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: [1]
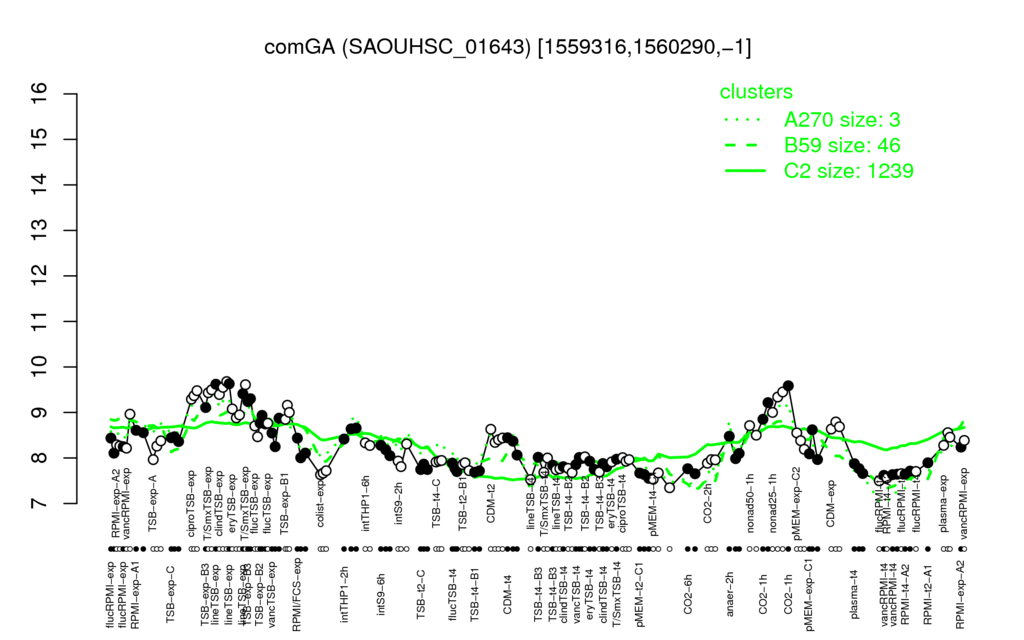 Multi-gene expression profiles
Multi-gene expression profiles
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
- Aureolib: no data available
⊟Protein stability[edit | edit source]
- half-life: no data available
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You can add further information about the gene and protein here. [edit]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ Ulrike Mäder, Pierre Nicolas, Maren Depke, Jan Pané-Farré, Michel Debarbouille, Magdalena M van der Kooi-Pol, Cyprien Guérin, Sandra Dérozier, Aurelia Hiron, Hanne Jarmer, Aurélie Leduc, Stephan Michalik, Ewoud Reilman, Marc Schaffer, Frank Schmidt, Philippe Bessières, Philippe Noirot, Michael Hecker, Tarek Msadek, Uwe Völker, Jan Maarten van Dijl
Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions.
PLoS Genet: 2016, 12(4);e1005962
[PubMed:27035918] [WorldCat.org] [DOI] (I e)
