Jump to navigation
Jump to search
PangenomeCOLN315NCTC8325NewmanUSA300_FPR3757JSNZ04-0298108BA0217611819-97685071193ECT-R 2ED133ED98HO 5096 0412JH1JH9JKD6008JKD6159LGA251M013MRSA252MSHR1132MSSA476MW2Mu3Mu50RF122ST398T0131TCH60TW20USA300_TCH1516VC40
NCBI: 03-AUG-2016
⊟Summary[edit | edit source]
- organism: Staphylococcus aureus NCTC8325
- locus tag: SAOUHSC_01535
- pan locus tag?: SAUPAN001831000
- symbol: SAOUHSC_01535
- pan gene symbol?: —
- synonym:
- product: hypothetical protein
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
- type: CDS
- locus tag: SAOUHSC_01535
- symbol: SAOUHSC_01535
- product: hypothetical protein
- replicon: chromosome
- strand: -
- coordinates: 1482837..1484000
- length: 1164
- essential: no DEG
⊟Accession numbers[edit | edit source]
- Gene ID: 3920637 NCBI
- RefSeq: YP_500051 NCBI
- BioCyc: G1I0R-1425 BioCyc
- MicrobesOnline: 1289965 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421
481
541
601
661
721
781
841
901
961
1021
1081
1141ATGCCGACATTATATGAATTAAAACAATCCTTAGGTATGATTGGACAACAATTAAAAAAT
AAAAATGATGAATTGAGTCAGAAAGCAACAGATCCAAATATTGATATGGAAGACATCAAA
CAACTAGAAACAGAAAAAGCAGGTTTACAACAAAGATTTAACATTGTTGAAAGACAAGTG
CAAGACATTGAAGAAAAAGAAAAAGCGAAAGTTAAAGATAAAGGAGAAGCTTATCAATCT
TTAAGTGATAATGAGAAGATGGTTAAAGCTAAGGCAGAGTTTTATCGTCACGCGATTTTA
CCAAATGAATTTGAAAAACCTTCAATGGAGGCACAACGTTTATTACACGCTTTACCAACA
GGAAATGATTCAGGTGGAGATAAGCTCTTACCAAAAACACTTTCTAAAGAAATTGTTTCA
GAACCATTTGCTAAAAACCAATTACGTGAAAAAGCTCGTCTAACTAACATTAAAGGTTTA
GAGATTCCAAGAGTTTCATACACTTTAGACGATGATGATTTCATTACAGACGTAGAAACA
GCAAAAGAATTAAAAGCAAAAGGTGATACAGTCAAGTTCACTACTAATAAATTCAAAGTA
TTTGCTGCAATTTCAGATACTGTAATTCATGGATCAGATGTAGATTTAGTAAACTGGGTT
GAAAACGCACTACAATCAGGATTAGCAGCTAAAGAGCGTAAAGATGCCTTAGCAGTAAGT
CCTAAATCTGGATTAGAACACATGTCATTTTATAATGGATCTGTTAAAGAAGTTGAGGGA
GCAGACATGTATGATGCTATTATTAACGCTTTAGCAGATTTACATGAAGATTATCGTGAT
AACGCAACAATTTATATGCGATATGCAGATTATGTCAAAATTATTAGTGTTCTTTCAAAT
GGAACAACAAATTTCTTTGACACACCAGCAGAAAAAGTATTTGGCAAACCAGTAGTATTT
ACAGATGCAGCAGTTAAACCTATTGTGGGAGATTTCAATTATTTTGGAATTAACTATGAT
GGAACAACTTATGACACTGATAAAGATGTTAAAAAAGGCGAATATTTGTTTGTATTAACA
GCATGGTATGATCAGCAACGTACATTAGACAGTGCATTCAGAATTGCAAAAGCAAAAGAA
AATACAGGTCCATTACCCAGCTAA60
120
180
240
300
360
420
480
540
600
660
720
780
840
900
960
1020
1080
1140
1164
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SAOUHSC_01535
- symbol: SAOUHSC_01535
- description: hypothetical protein
- length: 387
- theoretical pI: 4.90802
- theoretical MW: 43500.7
- GRAVY: -0.564858
⊟Function[edit | edit source]
- TIGRFAM: Mobile and extrachromosomal element functions Prophage functions phage major capsid protein, HK97 family (TIGR01554; HMM-score: 98.7)
- TheSEED :
- Phage major capsid protein
- PFAM: Phage-coat (CL0373) Phage_capsid; Phage capsid family (PF05065; HMM-score: 59.5)and 4 moreno clan defined ATG16; Autophagy protein 16 (ATG16) (PF08614; HMM-score: 14.5)HAUS-augmin3; HAUS augmin-like complex subunit 3 (PF14932; HMM-score: 13.2)Cep57_CLD_2; Centrosome localisation domain of PPC89 (PF14197; HMM-score: 12.9)APG6_N; Apg6 coiled-coil region (PF17675; HMM-score: 9.7)
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors:
- effectors:
⊟Localization[edit | edit source]
- PSORTb: Cytoplasmic
- Cytoplasmic Score: 7.5
- Cytoplasmic Membrane Score: 1.15
- Cellwall Score: 0.62
- Extracellular Score: 0.73
- Internal Helices: 0
- DeepLocPro: Extracellular
- Cytoplasmic Score: 0.3477
- Cytoplasmic Membrane Score: 0.0004
- Cell wall & surface Score: 0.0052
- Extracellular Score: 0.6466
- LocateP: Intracellular
- Prediction by SwissProt Classification: Cytoplasmic
- Pathway Prediction: No pathway
- Intracellular possibility: 1
- Signal peptide possibility: -1
- N-terminally Anchored Score: 1
- Predicted Cleavage Site: No CleavageSite
- SignalP: no predicted signal peptide
- SP(Sec/SPI): 0.002766
- TAT(Tat/SPI): 0.000347
- LIPO(Sec/SPII): 0.000721
- predicted transmembrane helices (TMHMM): 0
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MPTLYELKQSLGMIGQQLKNKNDELSQKATDPNIDMEDIKQLETEKAGLQQRFNIVERQVQDIEEKEKAKVKDKGEAYQSLSDNEKMVKAKAEFYRHAILPNEFEKPSMEAQRLLHALPTGNDSGGDKLLPKTLSKEIVSEPFAKNQLREKARLTNIKGLEIPRVSYTLDDDDFITDVETAKELKAKGDTVKFTTNKFKVFAAISDTVIHGSDVDLVNWVENALQSGLAAKERKDALAVSPKSGLEHMSFYNGSVKEVEGADMYDAIINALADLHEDYRDNATIYMRYADYVKIISVLSNGTTNFFDTPAEKVFGKPVVFTDAAVKPIVGDFNYFGINYDGTTYDTDKDVKKGEYLFVLTAWYDQQRTLDSAFRIAKAKENTGPLPS
⊟Experimental data[edit | edit source]
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
⊟Regulation[edit | edit source]
- regulator:
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: [1]
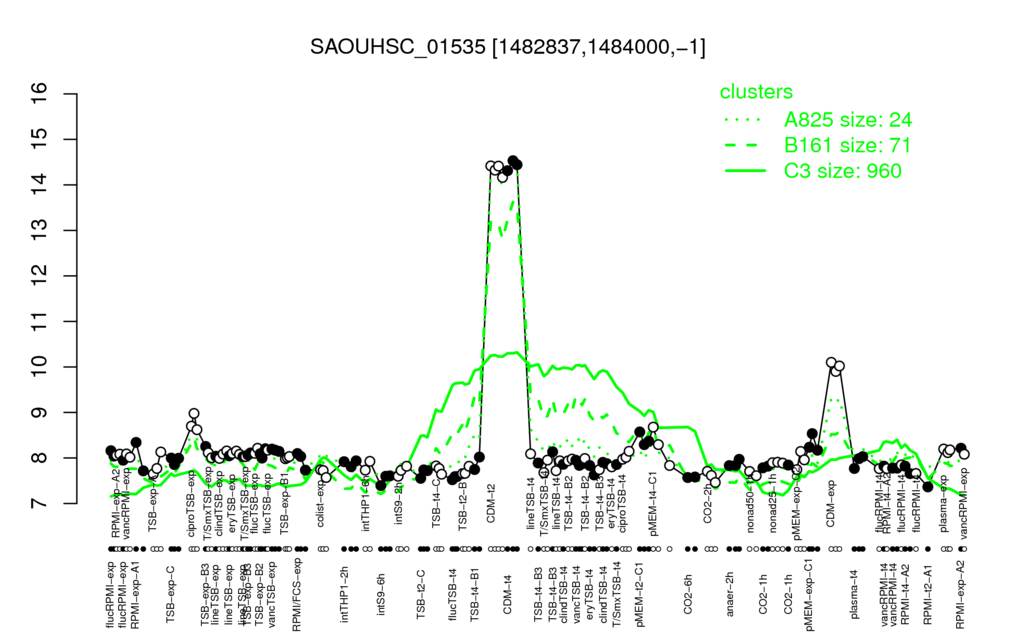 Multi-gene expression profiles
Multi-gene expression profiles
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
- Aureolib: no data available
⊟Protein stability[edit | edit source]
- half-life: no data available
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You can add further information about the gene and protein here. [edit]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ Ulrike Mäder, Pierre Nicolas, Maren Depke, Jan Pané-Farré, Michel Debarbouille, Magdalena M van der Kooi-Pol, Cyprien Guérin, Sandra Dérozier, Aurelia Hiron, Hanne Jarmer, Aurélie Leduc, Stephan Michalik, Ewoud Reilman, Marc Schaffer, Frank Schmidt, Philippe Bessières, Philippe Noirot, Michael Hecker, Tarek Msadek, Uwe Völker, Jan Maarten van Dijl
Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions.
PLoS Genet: 2016, 12(4);e1005962
[PubMed:27035918] [WorldCat.org] [DOI] (I e)
