Jump to navigation
Jump to search
PangenomeCOLN315NCTC8325NewmanUSA300_FPR3757JSNZ04-0298108BA0217611819-97685071193ECT-R 2ED133ED98HO 5096 0412JH1JH9JKD6008JKD6159LGA251M013MRSA252MSHR1132MSSA476MW2Mu3Mu50RF122ST398T0131TCH60TW20USA300_TCH1516VC40
NCBI: 03-AUG-2016
⊟Summary[edit | edit source]
- organism: Staphylococcus aureus NCTC8325
- locus tag: SAOUHSC_01536
- pan locus tag?: SAUPAN001832000
- symbol: SAOUHSC_01536
- pan gene symbol?: —
- synonym:
- product: scaffolding protease
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
- type: CDS
- locus tag: SAOUHSC_01536
- symbol: SAOUHSC_01536
- product: scaffolding protease
- replicon: chromosome
- strand: -
- coordinates: 1484012..1484785
- length: 774
- essential: no DEG
⊟Accession numbers[edit | edit source]
- Gene ID: 3920638 NCBI
- RefSeq: YP_500052 NCBI
- BioCyc: G1I0R-1426 BioCyc
- MicrobesOnline: 1289966 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421
481
541
601
661
721ATGTCAATGAAAGCTAAGTATTTTCAAATGAAAAGAAAATCAAAAAGTAAAGGTGAAATA
TTCATTTATGGTGATATTGTAAGTGATAAATGGTTTGAAAGTGATGTAACTGCTACAGAT
TTCAAAAATAAACTAGATGAACTAGGAGACATCAGTGAAATAGATGTTCATATAAATTCA
TCTGGAGGCAGTGTATTTGAAGGACATGCAATATACAATATGCTAAAAATGCATCCTGCA
AAAATTAATATCTATGTCGATGCCTTAGCGGCATCAATTGCTAGTGTTATCGCTATGAGT
GGTGACACTATTTTTATGCACAAAAATAGTTTTTTAATGATTCATAATTCATGGGTTATG
ACTGTAGGTAATGCAGAAGAATTAAGAAAGACAGCGGATTTACTTGAAAAAACAGATGCT
GTTAGTAATTCAGCTTATTTAGATAAAGCAAAAGATTTAGATCAAGAACACTTAAAACAG
ATGTTAGATGCAGAAACTTGGCTTACTGCAGAAGAAGCCTTGTCTTTCGGCTTGATAGAT
GAAATTTTAGGAGCTAATGAAATAGCTGCTAGTATCTCTAAAGAGCAATATAAGCGTTTC
GAGAACGTCCCAGAAGATTTAAAGAAAGATGTAGACAAAATCACTAAAATTGATGATGTA
GATACATCTGAATTGGTTGAAACACCTAAAGAAAGTATGTCACTAGAAGAAAAAGAAAAA
AGAGAAAAAATTAAACGCGAATGCGAAATTTTAAAAATGACAATGAATTATTAG60
120
180
240
300
360
420
480
540
600
660
720
774
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SAOUHSC_01536
- symbol: SAOUHSC_01536
- description: scaffolding protease
- length: 257
- theoretical pI: 4.71323
- theoretical MW: 29170.1
- GRAVY: -0.42179
⊟Function[edit | edit source]
- TIGRFAM: Protein fate Degradation of proteins, peptides, and glycopeptides ATP-dependent Clp endopeptidase, proteolytic subunit ClpP (TIGR00493; EC 3.4.21.92; HMM-score: 51.8)and 1 moreProtein fate Degradation of proteins, peptides, and glycopeptides signal peptide peptidase SppA, 36K type (TIGR00706; EC 3.4.-.-; HMM-score: 35.8)
- TheSEED :
- Phage head, head-tail preconnector protease C
- PFAM: ClpP_crotonase (CL0127) CLP_protease; Clp protease (PF00574; HMM-score: 104.1)and 4 moreSDH_protease; ClpP-like SDH-type serine proteinase (PF01972; HMM-score: 26.3)NfeD1b_N; NfeD1b, N-terminal domain (PF25145; HMM-score: 21.8)Peptidase_S49; Peptidase family S49 (PF01343; HMM-score: 18.1)COMMD_HN (CL0813) COMM_HN; COMMD2-7/10, HN domain (PF21672; HMM-score: 10)
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors:
- effectors:
⊟Localization[edit | edit source]
- PSORTb: unknown (no significant prediction)
- Cytoplasmic Score: 2.5
- Cytoplasmic Membrane Score: 2.5
- Cellwall Score: 2.5
- Extracellular Score: 2.5
- Internal Helices: 0
- DeepLocPro: Cytoplasmic
- Cytoplasmic Score: 0.5629
- Cytoplasmic Membrane Score: 0.14
- Cell wall & surface Score: 0.006
- Extracellular Score: 0.2911
- LocateP: Intracellular /TMH start AFTER 60
- Prediction by SwissProt Classification: Cytoplasmic
- Pathway Prediction: Possibly Sec-
- Intracellular possibility: 0.17
- Signal peptide possibility: -1
- N-terminally Anchored Score: 1
- Predicted Cleavage Site: No CleavageSite
- SignalP: no predicted signal peptide
- SP(Sec/SPI): 0.005815
- TAT(Tat/SPI): 0.000242
- LIPO(Sec/SPII): 0.000638
- predicted transmembrane helices (TMHMM): 1
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MSMKAKYFQMKRKSKSKGEIFIYGDIVSDKWFESDVTATDFKNKLDELGDISEIDVHINSSGGSVFEGHAIYNMLKMHPAKINIYVDALAASIASVIAMSGDTIFMHKNSFLMIHNSWVMTVGNAEELRKTADLLEKTDAVSNSAYLDKAKDLDQEHLKQMLDAETWLTAEEALSFGLIDEILGANEIAASISKEQYKRFENVPEDLKKDVDKITKIDDVDTSELVETPKESMSLEEKEKREKIKRECEILKMTMNY
⊟Experimental data[edit | edit source]
- experimentally validated: data available for COL
- protein localization:
- quantitative data / protein copy number per cell:
- interaction partners:
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
⊟Regulation[edit | edit source]
- regulator:
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: [1]
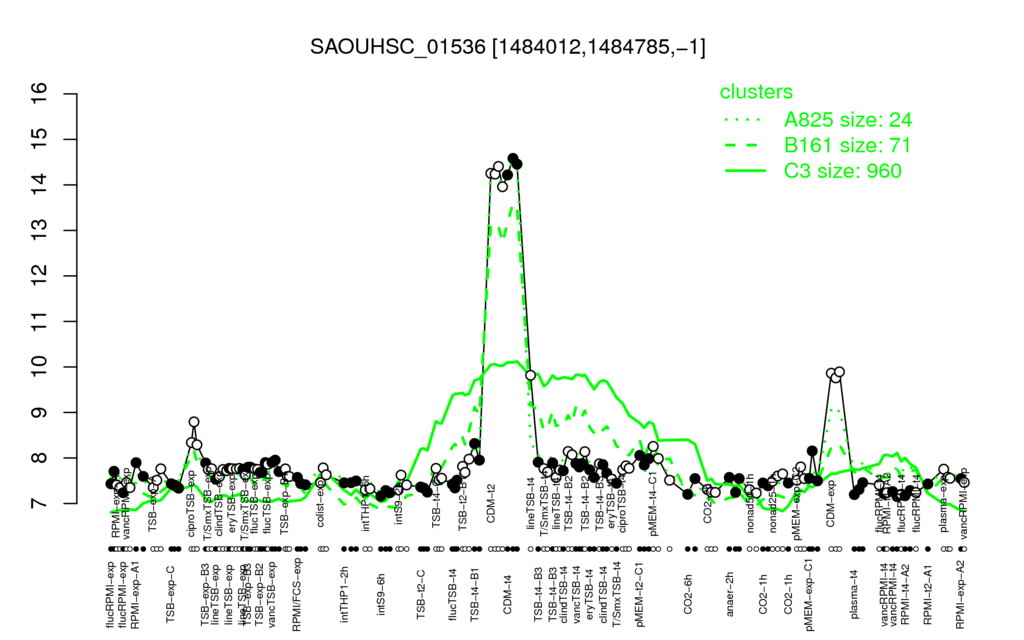 Multi-gene expression profiles
Multi-gene expression profiles
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
- Aureolib: no data available
⊟Protein stability[edit | edit source]
- half-life: no data available
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You can add further information about the gene and protein here. [edit]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ Ulrike Mäder, Pierre Nicolas, Maren Depke, Jan Pané-Farré, Michel Debarbouille, Magdalena M van der Kooi-Pol, Cyprien Guérin, Sandra Dérozier, Aurelia Hiron, Hanne Jarmer, Aurélie Leduc, Stephan Michalik, Ewoud Reilman, Marc Schaffer, Frank Schmidt, Philippe Bessières, Philippe Noirot, Michael Hecker, Tarek Msadek, Uwe Völker, Jan Maarten van Dijl
Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions.
PLoS Genet: 2016, 12(4);e1005962
[PubMed:27035918] [WorldCat.org] [DOI] (I e)
