Jump to navigation
Jump to search
PangenomeCOLN315NCTC8325NewmanUSA300_FPR3757JSNZ04-0298108BA0217611819-97685071193ECT-R 2ED133ED98HO 5096 0412JH1JH9JKD6008JKD6159LGA251M013MRSA252MSHR1132MSSA476MW2Mu3Mu50RF122ST398T0131TCH60TW20USA300_TCH1516VC40
NCBI: 03-AUG-2016
⊟Summary[edit | edit source]
- organism: Staphylococcus aureus NCTC8325
- locus tag: SAOUHSC_01538
- pan locus tag?: SAUPAN001834000
- symbol: SAOUHSC_01538
- pan gene symbol?: —
- synonym:
- product: phage terminase large subunit
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
- type: CDS
- locus tag: SAOUHSC_01538
- symbol: SAOUHSC_01538
- product: phage terminase large subunit
- replicon: chromosome
- strand: -
- coordinates: 1486012..1487703
- length: 1692
- essential: no DEG
⊟Accession numbers[edit | edit source]
- Gene ID: 3920581 NCBI
- RefSeq: YP_500054 NCBI
- BioCyc: G1I0R-1428 BioCyc
- MicrobesOnline: 1289968 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421
481
541
601
661
721
781
841
901
961
1021
1081
1141
1201
1261
1321
1381
1441
1501
1561
1621
1681GTGACTATTAAAGTTTTAAATGAACCTTCACCAAAACTATTAACAACATGGTATGCAGAG
CAAGTCACTCAAGGGAAAATAAAAACAAGCAAATATGTTAAAAAAGAATGTGAGAGACAC
CTTAGATATCTAGAAAATGGAGGTAAATGGGTATTTGATGAAGAATTAGCGCACCGTCCT
ATTCGATTCATAGAAAAGTTTTGTAAACCTTCCAAAGGATCTAAACGTCAACTTGTATTA
CAACCATGGCAACATTTTATTATTGGCAGTTTGTTTGGTTGGGTTCATAAAGAAACAAAA
CTGCGCAGGTTTAAAGAAGCTTTGATATTTATGGGGCGAAAAAATGGTAAAACAACTACT
ATATCTGGTGTTGCTAACTATGCTGTTTCTCAAGATGGAGAAAACGGCGCTGAAATCCAT
CTTTTAGCAAACGTAATGAAACAAGCTAGGATTCTATTTGATGAATCTAAGGCGATGATT
AAAGCTAGCCCAAAGCTTAGAGAAAATTTTAGACCTTTGAGAGATGAAATTCATTACGAT
GCAACTATATCTAAAATTATGCCACAGGCTTCAGACAGTGATAAGTTGGATGGTTTAAAT
ACACATATGGGCATTTTTGATGAAATTCATGAATTTAAAGATTATAAATTGATTTCAGTT
ATAAAAAACTCAAGAGCGGCAAGGTTACAACCCCTTCTTATCTACATTACGACAGCAGGG
TACCAACTAGATGGACCACTTGTTAATATGGTAGAAGCGGGAAGAGACACCTTAGATCGA
ATCATCGAAGATGAAAGAACTTTTTACTATTTAGCTTCTCTCGATGATGACGATGATATA
AATGATTCGTCGAATTGGATTAAAGCAAATCCTAACCTAGGTGTTTCTATCGATTTAGCT
GAAATGAAAGAAGAGTGGGAAAAGGCTAAGAGAACACCAGATGAACGTGGAGATTTTATA
ACCAAAAGGTTTAACATCTTTGCTAATAATGATGAGATGAGTTTTATTGATTATCCAACA
CTTCAAAAAAATAATGACATTATTTCCTTAGATGAGTTGGAAGGTAGACCATGTACTATA
GGTTATGATTTATCAGAAACAGAGGACTTTACAGCCGCATGTGCCACTTTTGCATTAGAT
AATGGCAAAGTTGCTGTCTTAACACATTCTTGGATTCCTAAGCATAAAGTTGAATATTCT
AACGAAAAGATACCCTATATAGAATGGGAAGAAGACGGATTACTAACAATACAAGATAAT
CCTTATATAGACTACCAAGATGTTTTAAATTGGATAATAAAGATGAATGAGCATTATGTT
GTCGAAAAAATCACTTATGATAGGGCGAATGCTTTTAAATTAAATCAAGAGTTAAAGAAT
TATGGCTTTGAAACAGAAGAAACAAGACAAGGGGCTTTGACCTTGAGCCCTGCATTGAAG
GATCTAAAAGAAATGTTTTTAGATGGGAAAATAATATTTAATAATAATCCTTTAATGAAA
TGGTATATCAATAATGTTCAGCTGAAACTAGACAGAAATGGGAACTGGCTGCCATCTAAA
CAAAGCAGATATCGTAAAATAGATGGTTTTGCAGCATTTTTAAACACATATACAGATATT
ATGAATAAAGTTGTTTCTGACAAGGGTGAAGGAAACATAGAATTTATTAGTATTAAAGAT
ATAATGCGTTAA60
120
180
240
300
360
420
480
540
600
660
720
780
840
900
960
1020
1080
1140
1200
1260
1320
1380
1440
1500
1560
1620
1680
1692
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SAOUHSC_01538
- symbol: SAOUHSC_01538
- description: phage terminase large subunit
- length: 563
- theoretical pI: 6.0331
- theoretical MW: 65246.9
- GRAVY: -0.54103
⊟Function[edit | edit source]
- TIGRFAM:
- TheSEED :
- Phage terminase large subunit
- PFAM: RNase_H (CL0219) TerL_nuclease; Terminase large subunit, endonuclease domain (PF20441; HMM-score: 149.4)and 5 moreP-loop_NTPase (CL0023) TerL_ATPase; Terminase large subunit, ATPase domain (PF03354; HMM-score: 101.2)no clan defined CagD; Pathogenicity island component CagD (PF16567; HMM-score: 13.2)COP23; Circadian oscillating protein COP23 (PF14218; HMM-score: 13)Tail_tube; Phage tail tube protein (PF10618; HMM-score: 12.2)P-loop_NTPase (CL0023) GpA_ATPase; Phage terminase large subunit gpA, ATPase domain (PF05876; HMM-score: 11.8)
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors:
- effectors:
⊟Localization[edit | edit source]
- PSORTb: Cytoplasmic
- Cytoplasmic Score: 7.5
- Cytoplasmic Membrane Score: 1.15
- Cellwall Score: 0.62
- Extracellular Score: 0.73
- Internal Helices: 0
- DeepLocPro: Cytoplasmic
- Cytoplasmic Score: 0.9473
- Cytoplasmic Membrane Score: 0.0156
- Cell wall & surface Score: 0.0004
- Extracellular Score: 0.0368
- LocateP: Intracellular
- Prediction by SwissProt Classification: Cytoplasmic
- Pathway Prediction: No pathway
- Intracellular possibility: 1
- Signal peptide possibility: -1
- N-terminally Anchored Score: 1
- Predicted Cleavage Site: No CleavageSite
- SignalP: no predicted signal peptide
- SP(Sec/SPI): 0.011278
- TAT(Tat/SPI): 0.000308
- LIPO(Sec/SPII): 0.003165
- predicted transmembrane helices (TMHMM): 0
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MTIKVLNEPSPKLLTTWYAEQVTQGKIKTSKYVKKECERHLRYLENGGKWVFDEELAHRPIRFIEKFCKPSKGSKRQLVLQPWQHFIIGSLFGWVHKETKLRRFKEALIFMGRKNGKTTTISGVANYAVSQDGENGAEIHLLANVMKQARILFDESKAMIKASPKLRENFRPLRDEIHYDATISKIMPQASDSDKLDGLNTHMGIFDEIHEFKDYKLISVIKNSRAARLQPLLIYITTAGYQLDGPLVNMVEAGRDTLDRIIEDERTFYYLASLDDDDDINDSSNWIKANPNLGVSIDLAEMKEEWEKAKRTPDERGDFITKRFNIFANNDEMSFIDYPTLQKNNDIISLDELEGRPCTIGYDLSETEDFTAACATFALDNGKVAVLTHSWIPKHKVEYSNEKIPYIEWEEDGLLTIQDNPYIDYQDVLNWIIKMNEHYVVEKITYDRANAFKLNQELKNYGFETEETRQGALTLSPALKDLKEMFLDGKIIFNNNPLMKWYINNVQLKLDRNGNWLPSKQSRYRKIDGFAAFLNTYTDIMNKVVSDKGEGNIEFISIKDIMR
⊟Experimental data[edit | edit source]
- experimentally validated:
- protein localization:
- quantitative data / protein copy number per cell:
- interaction partners:
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
⊟Regulation[edit | edit source]
- regulator:
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: [1]
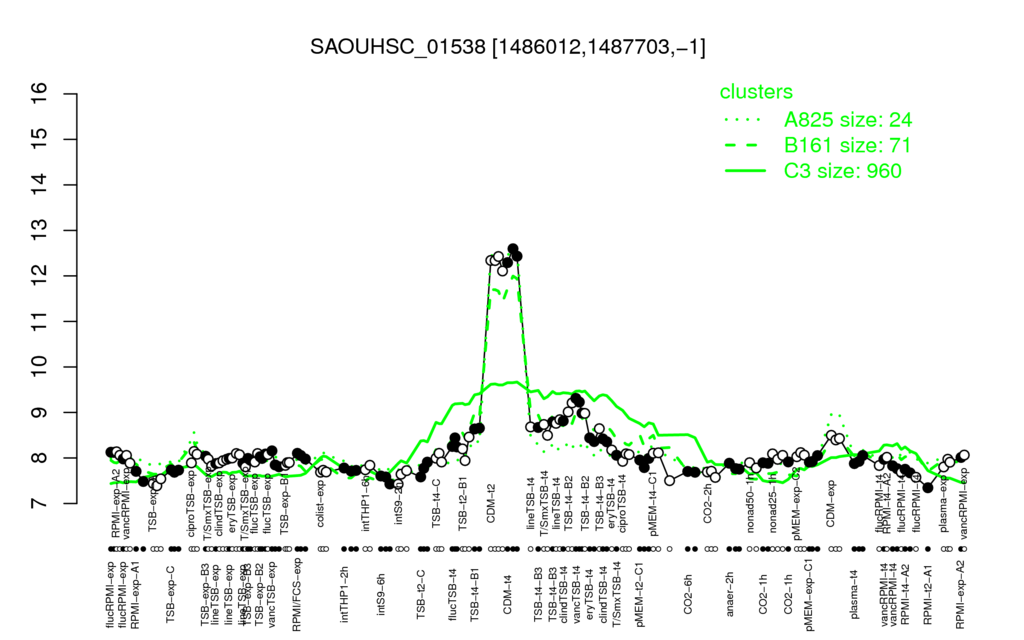 Multi-gene expression profiles
Multi-gene expression profiles
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
- Aureolib: no data available
⊟Protein stability[edit | edit source]
- half-life: no data available
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You can add further information about the gene and protein here. [edit]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ Ulrike Mäder, Pierre Nicolas, Maren Depke, Jan Pané-Farré, Michel Debarbouille, Magdalena M van der Kooi-Pol, Cyprien Guérin, Sandra Dérozier, Aurelia Hiron, Hanne Jarmer, Aurélie Leduc, Stephan Michalik, Ewoud Reilman, Marc Schaffer, Frank Schmidt, Philippe Bessières, Philippe Noirot, Michael Hecker, Tarek Msadek, Uwe Völker, Jan Maarten van Dijl
Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions.
PLoS Genet: 2016, 12(4);e1005962
[PubMed:27035918] [WorldCat.org] [DOI] (I e)
