⊟Summary[edit | edit source]
- organism: Staphylococcus aureus NCTC8325
- locus tag: SAOUHSC_02635
- pan locus tag?: SAUPAN005861000
- symbol: SAOUHSC_02635
- pan gene symbol?: tcaA
- synonym:
- product: hypothetical protein
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
- type: CDS
- locus tag: SAOUHSC_02635
- symbol: SAOUHSC_02635
- product: hypothetical protein
- replicon: chromosome
- strand: -
- coordinates: 2421820..2423202
- length: 1383
- essential: no DEG other strains
⊟Accession numbers[edit | edit source]
- Gene ID: 3921412 NCBI
- RefSeq: YP_501096 NCBI
- BioCyc: G1I0R-2485 BioCyc
- MicrobesOnline: 1291067 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421
481
541
601
661
721
781
841
901
961
1021
1081
1141
1201
1261
1321
1381ATGAAATCTTGCCCGAAGTGCGGACAACAAGCACAAGATGATGTACAGATATGTACACAA
TGTGGACATAAATTTGATAGTCGTCAAGCATTATATAGAAAGTCAACCGATGAAGATATA
CAAACTAATAACATCAAGATGAGAAAAATGGTGCCTTGGGCAATTGGTTTTTTCATTTTA
ATCTTAATTATTATATTGTTCTTTTTACTAAGAAACTTCAATTCACCTGAAGCGCAAACT
AAAATATTAGTGAATGCGATTGAGAATAATGATAAACAAAAAGTAGCAACATTATTAAGT
ACTAAAGATAATAAAGTAGATTCTGAAGAAGCAAAAGTATACATTAACTATATCAAAGAT
GAAGTTGGGCTTAAGCAATTTGTCAGCGACCTTAAAAATACGGTACATAAATTGAATAAG
AGTAAGACCAGCGTAGCTTCTTATATTCAAACCAGATCTGGTCAAAATATATTACGTGTA
AGTAAAAATGGCACACGTTATATCTTTTTCGATAATATGAGCTTTACTGCACCTACCAAG
CAACCAATTGTTAAACCGAAAGAAAAAACAAAATATGAGTTTAAATCTGGTGGTAAGAAA
AAGATGGTTATAGCTGAAGCAAATAAAGTGACGCCAATAGGTAATTTTATACCGGGGACA
TATAGAATTCCAGCTATGAAATCAACTGAGAACGGTGATTTTGCAGGCCATTTAAAATTT
GATTTTAGACAAAGTAATTCTGAAACGGTAGATGTTACTGAAGATTTTGAAGAAGCAAAT
ATATCTGTAACTTTAAAAGGCGATACAAAATTAAATGATAGTTCTAAAAAAGTAACTATA
AATGACCATGAAATGGCATTTTCAAGTTCCAAAACGTATGGTCCATATCCACAAAATAAA
GATATTACCATTTCAGCTTCAGGTAAAGCGAAAGATAAAACATTTACAACACAGACGAAG
ACGATTAAAGCTAGCGATTTAAAATACAATACAGAGATAACTTTGAATTTTGACAGTGAA
GATATCGAAGACTATGTTGAAAAGAAAGAAAAAGAAGAAAACAGCTTGAAGAACAAATTG
ATAGAATTCTTTGCTGGATATTCTTTAGCGAATAATGCTGCGTTTAATCAGTCGGATTTT
GATTTTGTATCATCATATATAAAAAAAGGATCATCTTTTTATGATGATGTAAAGAAGCGT
GTATCTAAAGGAAGTTTAATGATGATTAGTTCACCACAAATTATAGATGCTGAAAAACAT
GGTGATAAGATTACCGCAACCGTAAGATTAATAAATGAAAACGGTAAGCAAGTAGATAAA
GAATATGAGCTTGAACAAGGCTCGCAAGACCGCTTGCAATTAATCAAGACATCAGAAAAA
TAG60
120
180
240
300
360
420
480
540
600
660
720
780
840
900
960
1020
1080
1140
1200
1260
1320
1380
1383
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SAOUHSC_02635
- symbol: SAOUHSC_02635
- description: hypothetical protein
- length: 460
- theoretical pI: 9.47124
- theoretical MW: 52113.8
- GRAVY: -0.652826
⊟Function[edit | edit source]
- TIGRFAM: Regulatory functions DNA interactions putative regulatory protein, FmdB family (TIGR02605; HMM-score: 13.7)and 1 moreCys-rich peptide, TIGR04165 family (TIGR04165; HMM-score: 5.7)
- TheSEED :
- Teicoplanin resistance associated membrane protein TcaA
- PFAM: NTF2 (CL0051) TcaA_5th; TcaA protein NTF2-like domain (PF22819; HMM-score: 82.5)no clan defined TcaA_2nd; TcaA protein second domain (PF22813; HMM-score: 68.8)and 19 moreE-set (CL0159) TcaA_3rd_4th; TcaA protein 3rd/4th domain (PF22820; HMM-score: 34.7)Zn_Beta_Ribbon (CL0167) Zn_ribbon_3; zinc-ribbon domain (PF13248; HMM-score: 27.7)Zn_Ribbon_1; zinc-ribbon domain (PF13240; HMM-score: 25.9)UPF0547; Uncharacterised protein family UPF0547 (PF10571; HMM-score: 19.8)DZR; Double zinc ribbon (PF12773; HMM-score: 18.1)no clan defined DUF2950; Protein of unknown function (DUF2950) (PF11453; HMM-score: 15)EssA; WXG100 protein secretion system (Wss), protein EssA (PF10661; HMM-score: 14.7)Zn_Beta_Ribbon (CL0167) Zn_ribbon_8; Zinc ribbon domain (PF09723; HMM-score: 13.9)ClpP_crotonase (CL0127) CLP_protease; Clp protease (PF00574; HMM-score: 13.4)MBB (CL0193) OprB; Carbohydrate-selective porin, OprB family (PF04966; HMM-score: 12.9)Zn_Beta_Ribbon (CL0167) DZR_2; Double zinc ribbon domain (PF18912; HMM-score: 12.5)DUF7575; Domain of unknown function (DUF7575) (PF24460; HMM-score: 12.4)ABC-2 (CL0181) ABC2_membrane_3; ABC-2 family transporter protein (PF12698; HMM-score: 11.5)Zn_Beta_Ribbon (CL0167) Zn_ribbon_15; zinc-ribbon family (PF17032; HMM-score: 11.3)Cas12f1-like_TNB; Cas12f1-like, TNB domain (PF07282; HMM-score: 10.8)RING (CL0229) Prok-RING_1; Prokaryotic RING finger family 1 (PF14446; HMM-score: 10.4)Transporter (CL0375) Clc-like; Clc-like (PF07062; HMM-score: 9.9)no clan defined DUF7765; Domain of unknown function (DUF7765) (PF24958; HMM-score: 7.6)YhfH; YhfH-like protein (PF14149; HMM-score: 7.2)
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors:
- effectors:
⊟Localization[edit | edit source]
- PSORTb: unknown (no significant prediction)
- Cytoplasmic Score: 2.5
- Cytoplasmic Membrane Score: 2.5
- Cellwall Score: 2.5
- Extracellular Score: 2.5
- Internal Helix: 1
- DeepLocPro: Cytoplasmic Membrane
- Cytoplasmic Score: 0.0001
- Cytoplasmic Membrane Score: 0.8618
- Cell wall & surface Score: 0.0015
- Extracellular Score: 0.1366
- LocateP: N-terminally anchored (No CS)
- Prediction by SwissProt Classification: Membrane
- Pathway Prediction: Sec-(SPI)
- Intracellular possibility: 0.17
- Signal peptide possibility: -1
- N-terminally Anchored Score: 4
- Predicted Cleavage Site: No CleavageSite
- SignalP: no predicted signal peptide
- SP(Sec/SPI): 0.008059
- TAT(Tat/SPI): 0.000171
- LIPO(Sec/SPII): 0.029427
- predicted transmembrane helices (TMHMM): 1
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MKSCPKCGQQAQDDVQICTQCGHKFDSRQALYRKSTDEDIQTNNIKMRKMVPWAIGFFILILIIILFFLLRNFNSPEAQTKILVNAIENNDKQKVATLLSTKDNKVDSEEAKVYINYIKDEVGLKQFVSDLKNTVHKLNKSKTSVASYIQTRSGQNILRVSKNGTRYIFFDNMSFTAPTKQPIVKPKEKTKYEFKSGGKKKMVIAEANKVTPIGNFIPGTYRIPAMKSTENGDFAGHLKFDFRQSNSETVDVTEDFEEANISVTLKGDTKLNDSSKKVTINDHEMAFSSSKTYGPYPQNKDITISASGKAKDKTFTTQTKTIKASDLKYNTEITLNFDSEDIEDYVEKKEKEENSLKNKLIEFFAGYSLANNAAFNQSDFDFVSSYIKKGSSFYDDVKKRVSKGSLMMISSPQIIDAEKHGDKITATVRLINENGKQVDKEYELEQGSQDRLQLIKTSEK
⊟Experimental data[edit | edit source]
- experimentally validated: PeptideAtlas [1] [2]
- protein localization: data available for COL
- quantitative data / protein copy number per cell:
- interaction partners:
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
- predicted SigA promoter [3] : SAOUHSC_02635 < S1021 < SAOUHSC_02636 < S1022 < S1023
⊟Regulation[edit | edit source]
- regulator: VraR* (activation) regulon
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: [3]
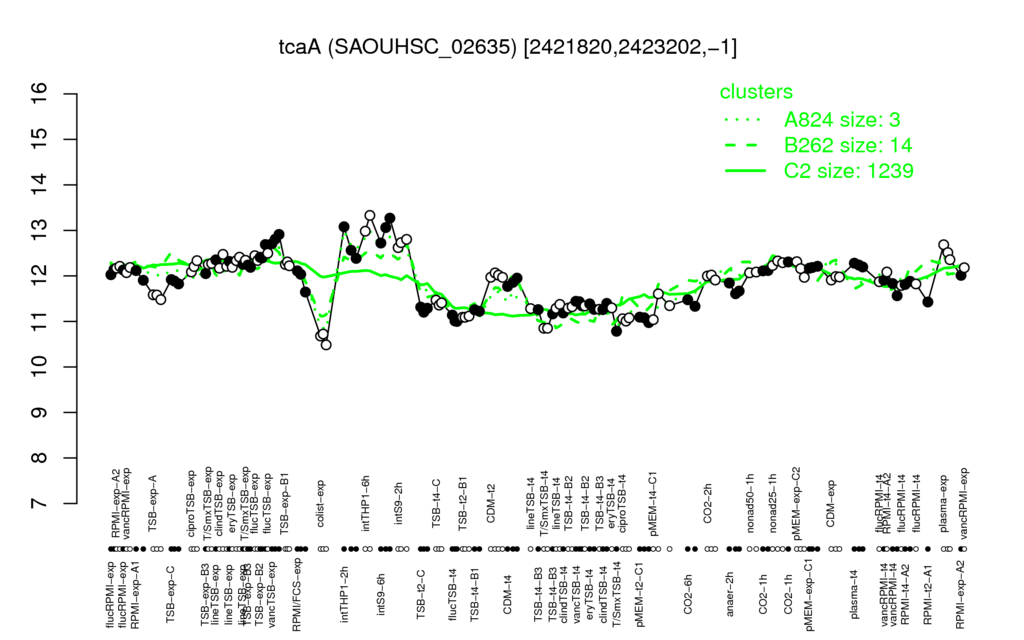 Multi-gene expression profiles
Multi-gene expression profiles
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
- Aureolib: no data available
⊟Protein stability[edit | edit source]
- half-life: no data available
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You can add further information about the gene and protein here. [edit]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ Maren Depke, Stephan Michalik, Alexander Rabe, Kristin Surmann, Lars Brinkmann, Nico Jehmlich, Jörg Bernhardt, Michael Hecker, Bernd Wollscheid, Zhi Sun, Robert L Moritz, Uwe Völker, Frank Schmidt
A peptide resource for the analysis of Staphylococcus aureus in host-pathogen interaction studies.
Proteomics: 2015, 15(21);3648-61
[PubMed:26224020] [WorldCat.org] [DOI] (I p) - ↑ Stephan Michalik, Maren Depke, Annette Murr, Manuela Gesell Salazar, Ulrike Kusebauch, Zhi Sun, Tanja C Meyer, Kristin Surmann, Henrike Pförtner, Petra Hildebrandt, Stefan Weiss, Laura Marcela Palma Medina, Melanie Gutjahr, Elke Hammer, Dörte Becher, Thomas Pribyl, Sven Hammerschmidt, Eric W Deutsch, Samuel L Bader, Michael Hecker, Robert L Moritz, Ulrike Mäder, Uwe Völker, Frank Schmidt
A global Staphylococcus aureus proteome resource applied to the in vivo characterization of host-pathogen interactions.
Sci Rep: 2017, 7(1);9718
[PubMed:28887440] [WorldCat.org] [DOI] (I e) - ↑ 3.0 3.1 3.2 Ulrike Mäder, Pierre Nicolas, Maren Depke, Jan Pané-Farré, Michel Debarbouille, Magdalena M van der Kooi-Pol, Cyprien Guérin, Sandra Dérozier, Aurelia Hiron, Hanne Jarmer, Aurélie Leduc, Stephan Michalik, Ewoud Reilman, Marc Schaffer, Frank Schmidt, Philippe Bessières, Philippe Noirot, Michael Hecker, Tarek Msadek, Uwe Völker, Jan Maarten van Dijl
Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions.
PLoS Genet: 2016, 12(4);e1005962
[PubMed:27035918] [WorldCat.org] [DOI] (I e) - ↑ Makoto Kuroda, Hiroko Kuroda, Taku Oshima, Fumihiko Takeuchi, Hirotada Mori, Keiichi Hiramatsu
Two-component system VraSR positively modulates the regulation of cell-wall biosynthesis pathway in Staphylococcus aureus.
Mol Microbiol: 2003, 49(3);807-21
[PubMed:12864861] [WorldCat.org] [DOI] (P p) - ↑ Susan Boyle-Vavra, Shouhui Yin, Dae Sun Jo, Christopher P Montgomery, Robert S Daum
VraT/YvqF is required for methicillin resistance and activation of the VraSR regulon in Staphylococcus aureus.
Antimicrob Agents Chemother: 2013, 57(1);83-95
[PubMed:23070169] [WorldCat.org] [DOI] (I p)
⊟Relevant publications[edit | edit source]
M Brandenberger, M Tschierske, P Giachino, A Wada, B Berger-Bächi
Inactivation of a novel three-cistronic operon tcaR-tcaA-tcaB increases teicoplanin resistance in Staphylococcus aureus.
Biochim Biophys Acta: 2000, 1523(2-3);135-9
[PubMed:11042376] [WorldCat.org] [DOI] (P p)Hideki Maki, Nadine McCallum, Markus Bischoff, Akihito Wada, Brigitte Berger-Bächi
tcaA inactivation increases glycopeptide resistance in Staphylococcus aureus.
Antimicrob Agents Chemother: 2004, 48(6);1953-9
[PubMed:15155184] [WorldCat.org] [DOI] (P p)N McCallum, G Spehar, M Bischoff, B Berger-Bächi
Strain dependence of the cell wall-damage induced stimulon in Staphylococcus aureus.
Biochim Biophys Acta: 2006, 1760(10);1475-81
[PubMed:16891058] [WorldCat.org] [DOI] (P p)
