Jump to navigation
Jump to search
NCBI: 03-AUG-2016
⊟Summary[edit | edit source]
- organism: Staphylococcus aureus NCTC8325
- locus tag: SAOUHSC_01668
- pan locus tag?: SAUPAN004148000
- symbol: era
- pan gene symbol?: era
- synonym:
- product: GTP-binding protein Era
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
⊟Accession numbers[edit | edit source]
- Gene ID: 3920080 NCBI
- RefSeq: YP_500179 NCBI
- BioCyc: G1I0R-1551 BioCyc
- MicrobesOnline: 1290093 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421
481
541
601
661
721
781
841ATGACAGAACATAAATCAGGATTTGTTTCAATTATAGGTAGACCAAATGTAGGAAAGTCA
ACATTTGTTAATAGAGTGATCGGCCATAAAATAGCAATCATGTCCGATAAAGCTCAAACA
ACTAGAAATAAAATTCAAGGTGTTATGACAAGAGATGACGCGCAAATTATATTCATTGAT
ACGCCAGGTATTCATAAACCTAAACACAAATTAGGTGACTATATGATGAAAGTCGCTAAA
AATACATTATCTGAGATAGATGCAATCATGTTTATGGTTAATGCCAATGAGGAAATTGGA
CGAGGCGATGAATATATTATAGAAATGTTGAAAAATGTTAAGACACCAGTATTTTTAGTA
TTAAATAAAATAGATTTAGTGCATCCAGATGAATTAATGCCAAAGATTGAAGAATATCAA
AGTTATATGGACTTTACAGAGATTGTACCTATTTCAGCATTAGAAGGGCTAAATGTCGAT
CATTTTATTGATGTTTTAAAGACGTATTTACCCGAAGGACCTAAATATTATCCAGATGAT
CAAATTTCAGACCATCCTGAACAATTTGTAGTGGGTGAAATCATTCGTGAAAAAATCCTT
CATCTTACAAGTGAAGAAATCCCTCATGCGATTGGTGTTAATGTGGACCGTATGGTTAAA
GAAAGCGAAGATCGTGTTCATATCGAAGCAACTATATATGTTGAAAGAGGTTCGCAAAAA
GGAATTGTCATTGGAAAAGGCGGTAAAAAGTTAAAAGAAGTAGGAAAACGTGCGAGACGT
GATATAGAAATGCTTCTAGGCTCTAAAGTTTACTTAGAATTATGGGTCAAAGTTCAAAGA
GACTGGCGAAACAAAGTTAACTTTATTCGCCAAATTGGTTATGTTGAAGACCAAGATTAA60
120
180
240
300
360
420
480
540
600
660
720
780
840
900
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SAOUHSC_01668
- symbol: Era
- description: GTP-binding protein Era
- length: 299
- theoretical pI: 6.69067
- theoretical MW: 34270.4
- GRAVY: -0.343478
⊟Function[edit | edit source]
- TIGRFAM: Protein synthesis Other GTP-binding protein Era (TIGR00436; HMM-score: 350.3)and 18 moreProtein synthesis Other ribosome-associated GTPase EngA (TIGR03594; HMM-score: 133.5)Unknown function General small GTP-binding protein domain (TIGR00231; HMM-score: 96.3)Protein fate Protein modification and repair [FeFe] hydrogenase H-cluster maturation GTPase HydF (TIGR03918; HMM-score: 79.3)Protein synthesis tRNA and rRNA base modification tRNA modification GTPase TrmE (TIGR00450; EC 3.6.-.-; HMM-score: 71.3)Protein synthesis Other ribosome biogenesis GTP-binding protein YlqF (TIGR03596; HMM-score: 57.9)Protein synthesis Other ribosome biogenesis GTP-binding protein YsxC (TIGR03598; HMM-score: 54.2)Protein synthesis Other Obg family GTPase CgtA (TIGR02729; HMM-score: 41.3)Transport and binding proteins Cations and iron carrying compounds ferrous iron transport protein B (TIGR00437; HMM-score: 41.1)Unknown function General GTP-binding protein HflX (TIGR03156; HMM-score: 40)Protein synthesis Other ribosome biogenesis GTPase YqeH (TIGR03597; HMM-score: 37.4)Protein synthesis Translation factors translation initiation factor IF-2 (TIGR00487; HMM-score: 35.2)translation initiation factor 2, gamma subunit (TIGR03680; HMM-score: 34.1)Transport and binding proteins Nucleosides, purines and pyrimidines GTP-binding protein (TIGR00991; HMM-score: 20.7)Protein synthesis Translation factors translation initiation factor aIF-2 (TIGR00491; HMM-score: 18.5)Protein synthesis Translation factors ribosome small subunit-dependent GTPase A (TIGR00157; EC 3.6.-.-; HMM-score: 15.4)Transport and binding proteins Amino acids, peptides and amines chloroplast protein import component Toc86/159, G and M domains (TIGR00993; HMM-score: 15.4)nicotinamide-nucleotide adenylyltransferase (TIGR01526; EC 2.7.7.1; HMM-score: 13.5)Protein synthesis Translation factors peptide chain release factor 3 (TIGR00503; HMM-score: 13.4)
- TheSEED :
- GTP-binding protein Era
and 1 more - PFAM: P-loop_NTPase (CL0023) MMR_HSR1; 50S ribosome-binding GTPase (PF01926; HMM-score: 95.2)KH (CL0007) KH_2; KH domain (PF07650; HMM-score: 79.1)and 16 moreP-loop_NTPase (CL0023) GTP_EFTU; Elongation factor Tu GTP binding domain (PF00009; HMM-score: 53.9)FeoB_N; Ferrous iron transport protein B (PF02421; HMM-score: 49.6)Dynamin_N; Dynamin family (PF00350; HMM-score: 44.1)AIG1; AIG1 family (PF04548; HMM-score: 31.5)RsgA_GTPase; RsgA GTPase (PF03193; HMM-score: 30)cobW; CobW/HypB/UreG, nucleotide-binding domain (PF02492; HMM-score: 22)PduV-EutP; Ethanolamine utilisation - propanediol utilisation (PF10662; HMM-score: 21.8)no clan defined MMR_HSR1_Xtn; C-terminal region of MMR_HSR1 domain (PF16897; HMM-score: 20.4)P-loop_NTPase (CL0023) Ras; Ras family (PF00071; HMM-score: 19.1)Roc; Ras of Complex, Roc, domain of DAPkinase (PF08477; HMM-score: 17.9)AAA; ATPase family associated with various cellular activities (AAA) (PF00004; HMM-score: 15.3)ATP_bind_1; Conserved hypothetical ATP binding protein (PF03029; HMM-score: 14.3)nSTAND3; Novel STAND NTPase 3 (PF20720; HMM-score: 12.9)SpoIVA_ATPase; Stage IV sporulation protein A, ATPase domain (PF09547; HMM-score: 12.5)AAA_22; AAA domain (PF13401; HMM-score: 12.4)KH (CL0007) KH_KhpA-B; KhpA/KhpB, KH domain (PF13083; HMM-score: 11.9)
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors:
- effectors:
⊟Localization[edit | edit source]
- PSORTb: Cytoplasmic Membrane
- Cytoplasmic Score: 1.05
- Cytoplasmic Membrane Score: 8.78
- Cellwall Score: 0.08
- Extracellular Score: 0.09
- Internal Helices: 0
- DeepLocPro: Cytoplasmic
- Cytoplasmic Score: 0.9822
- Cytoplasmic Membrane Score: 0.014
- Cell wall & surface Score: 0
- Extracellular Score: 0.0037
- LocateP: Intracellular
- Prediction by SwissProt Classification: Cytoplasmic
- Pathway Prediction: No pathway
- Intracellular possibility: 1
- Signal peptide possibility: -1
- N-terminally Anchored Score: 1
- Predicted Cleavage Site: No CleavageSite
- SignalP: no predicted signal peptide
- SP(Sec/SPI): 0.003811
- TAT(Tat/SPI): 0.00048
- LIPO(Sec/SPII): 0.000587
- predicted transmembrane helices (TMHMM): 0
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MTEHKSGFVSIIGRPNVGKSTFVNRVIGHKIAIMSDKAQTTRNKIQGVMTRDDAQIIFIDTPGIHKPKHKLGDYMMKVAKNTLSEIDAIMFMVNANEEIGRGDEYIIEMLKNVKTPVFLVLNKIDLVHPDELMPKIEEYQSYMDFTEIVPISALEGLNVDHFIDVLKTYLPEGPKYYPDDQISDHPEQFVVGEIIREKILHLTSEEIPHAIGVNVDRMVKESEDRVHIEATIYVERGSQKGIVIGKGGKKLKEVGKRARRDIEMLLGSKVYLELWVKVQRDWRNKVNFIRQIGYVEDQD
⊟Experimental data[edit | edit source]
- experimentally validated: PeptideAtlas [2] [3]
- protein localization: data available for COL
- quantitative data / protein copy number per cell: data available for COL
- interaction partners:
SAOUHSC_01159 (ileS) isoleucyl-tRNA synthetase [4] (data from MRSA252) SAOUHSC_01243 (nusA) transcription elongation factor NusA [4] (data from MRSA252) SAOUHSC_00520 (rplJ) 50S ribosomal protein L10 [4] (data from MRSA252) SAOUHSC_02506 (rpsC) 30S ribosomal protein S3 [4] (data from MRSA252) SAOUHSC_00679 hypothetical protein [4] (data from MRSA252) SAOUHSC_00789 hypothetical protein [4] (data from MRSA252) SAOUHSC_00795 glyceraldehyde-3-phosphate dehydrogenase [4] (data from MRSA252) SAOUHSC_01028 phosphocarrier protein HPr [4] (data from MRSA252) SAOUHSC_01845 formate--tetrahydrofolate ligase [4] (data from MRSA252)
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
- MicrobesOnline: SAOUHSC_01667 < erapredicted SigA promoter [5] : SAOUHSC_01667 < era < SAOUHSC_01670 < SAOUHSC_01671 < SAOUHSC_01672 < SAOUHSC_01673 < S661
⊟Regulation[edit | edit source]
- regulator:
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: [5]
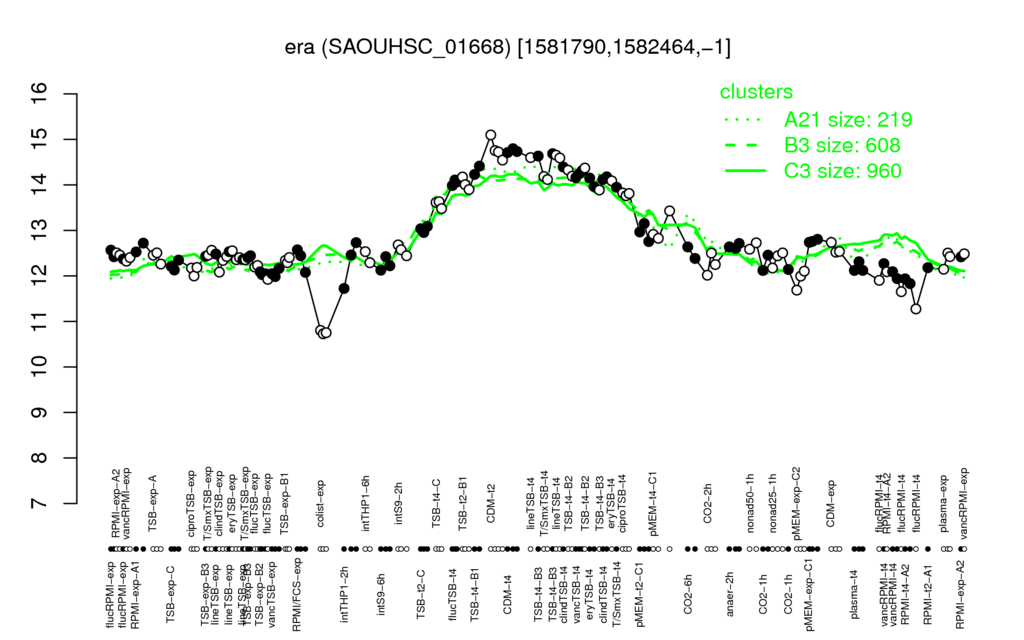 Multi-gene expression profiles
Multi-gene expression profiles
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
⊟Protein stability[edit | edit source]
- half-life: no data available
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You can add further information about the gene and protein here. [edit]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ Roy R Chaudhuri, Andrew G Allen, Paul J Owen, Gil Shalom, Karl Stone, Marcus Harrison, Timothy A Burgis, Michael Lockyer, Jorge Garcia-Lara, Simon J Foster, Stephen J Pleasance, Sarah E Peters, Duncan J Maskell, Ian G Charles
Comprehensive identification of essential Staphylococcus aureus genes using Transposon-Mediated Differential Hybridisation (TMDH).
BMC Genomics: 2009, 10;291
[PubMed:19570206] [WorldCat.org] [DOI] (I e) - ↑ Maren Depke, Stephan Michalik, Alexander Rabe, Kristin Surmann, Lars Brinkmann, Nico Jehmlich, Jörg Bernhardt, Michael Hecker, Bernd Wollscheid, Zhi Sun, Robert L Moritz, Uwe Völker, Frank Schmidt
A peptide resource for the analysis of Staphylococcus aureus in host-pathogen interaction studies.
Proteomics: 2015, 15(21);3648-61
[PubMed:26224020] [WorldCat.org] [DOI] (I p) - ↑ Stephan Michalik, Maren Depke, Annette Murr, Manuela Gesell Salazar, Ulrike Kusebauch, Zhi Sun, Tanja C Meyer, Kristin Surmann, Henrike Pförtner, Petra Hildebrandt, Stefan Weiss, Laura Marcela Palma Medina, Melanie Gutjahr, Elke Hammer, Dörte Becher, Thomas Pribyl, Sven Hammerschmidt, Eric W Deutsch, Samuel L Bader, Michael Hecker, Robert L Moritz, Ulrike Mäder, Uwe Völker, Frank Schmidt
A global Staphylococcus aureus proteome resource applied to the in vivo characterization of host-pathogen interactions.
Sci Rep: 2017, 7(1);9718
[PubMed:28887440] [WorldCat.org] [DOI] (I e) - ↑ 4.0 4.1 4.2 4.3 4.4 4.5 4.6 4.7 4.8 Artem Cherkasov, Michael Hsing, Roya Zoraghi, Leonard J Foster, Raymond H See, Nikolay Stoynov, Jihong Jiang, Sukhbir Kaur, Tian Lian, Linda Jackson, Huansheng Gong, Rick Swayze, Emily Amandoron, Farhad Hormozdiari, Phuong Dao, Cenk Sahinalp, Osvaldo Santos-Filho, Peter Axerio-Cilies, Kendall Byler, William R McMaster, Robert C Brunham, B Brett Finlay, Neil E Reiner
Mapping the protein interaction network in methicillin-resistant Staphylococcus aureus.
J Proteome Res: 2011, 10(3);1139-50
[PubMed:21166474] [WorldCat.org] [DOI] (I p) - ↑ 5.0 5.1 Ulrike Mäder, Pierre Nicolas, Maren Depke, Jan Pané-Farré, Michel Debarbouille, Magdalena M van der Kooi-Pol, Cyprien Guérin, Sandra Dérozier, Aurelia Hiron, Hanne Jarmer, Aurélie Leduc, Stephan Michalik, Ewoud Reilman, Marc Schaffer, Frank Schmidt, Philippe Bessières, Philippe Noirot, Michael Hecker, Tarek Msadek, Uwe Völker, Jan Maarten van Dijl
Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions.
PLoS Genet: 2016, 12(4);e1005962
[PubMed:27035918] [WorldCat.org] [DOI] (I e)
