Jump to navigation
Jump to search
NCBI: 03-AUG-2016
⊟Summary[edit | edit source]
- organism: Staphylococcus aureus NCTC8325
- locus tag: SAOUHSC_03012
- pan locus tag?: SAUPAN006432000
- symbol: SAOUHSC_03012
- pan gene symbol?: —
- synonym:
- product: histidinol-phosphate aminotransferase
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
- type: CDS
- locus tag: SAOUHSC_03012
- symbol: SAOUHSC_03012
- product: histidinol-phosphate aminotransferase
- replicon: chromosome
- strand: -
- coordinates: 2785375..2786388
- length: 1014
- essential: no DEG other strains
⊟Accession numbers[edit | edit source]
- Gene ID: 3921493 NCBI
- RefSeq: YP_501461 NCBI
- BioCyc: G1I0R-2833 BioCyc
- MicrobesOnline: 1291432 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421
481
541
601
661
721
781
841
901
961ATGATTTATATTGATAAAAATGAAAGTCCAGTTACGCCGTTGGATGAAAAAACAATGACG
TCTATTATTAGTGCAACGCCATATAATTTATATCCTGATGCAGCATATGAACAATTCAAG
GAAGCTTATGCTAAGTTTTACGGATTATCGCCTGAACAAATTATTGCAGGAAATGGATCT
GATGAATTGATTCAAAAGTTAATGCTGATCATGCCAGAAGGTCCGGCATTAACGCTAAAT
CCTGATTTTTTTATGTATCAAGCATATGCGGCACAAGTAAATCGTGAAATTGCATTTGTA
GATGCAGGATCAGATTTAACGTTTGATTTGGAAACCATTTTAACGAAAATCGATGAAGTA
CAACCATCATTTTTTATTATGAGTAATCCACATAACCCTTCAGGCAAGCAATTTGATACG
GCATTTTTAACAGCTATTGCAGATAAGATGAAAGCATTAAACGGATACTTTGTCATTGAT
GAAGCATATTTAGATTATGGTACGGCATATGACGTGGAACTGGCACCACACATCTTAAGA
ATGCGTACATTATCAAAGGCGTTTGGAATTGCCGGCTTAAGATTAGGTGTCTTAATTAGT
ACTGCTGGAACGATAAAGCATATTCAAAAAATAGAACATCCATATCCATTAAATGTATTT
ACGCTAAATATTGCGACTTATATTTTTAGACATAGAGAAGAGACAAGACAATTTTTAACG
ATGCAACGACAGTTAGCTGAGCAGTTAAAACAAATATTTGATACACATGTTGCAGATAAA
ATGTCAGTGTTCCCATCAAATGCTAATTTTGTACTTACTAAAGGCTCAGCAGCGCAACAA
TTAGGACAATACGTATATGAACAAGGATTTAAACCTCGCTTTTATGATGAGCCGGTGATG
AAGGGCTATGTAAGATACTCAATTGCAACAGCATCACAGTTAAAGCAATTAGAAGAAATT
GTTAAAGAATGGAGTGCAAAATATGATTTATCAAAAACAACGAAACACAGCTGA60
120
180
240
300
360
420
480
540
600
660
720
780
840
900
960
1014
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SAOUHSC_03012
- symbol: SAOUHSC_03012
- description: histidinol-phosphate aminotransferase
- length: 337
- theoretical pI: 5.65809
- theoretical MW: 38240.5
- GRAVY: -0.201187
⊟Function[edit | edit source]
- reaction: EC 2.6.1.9? ExPASyHistidinol-phosphate transaminase L-histidinol phosphate + 2-oxoglutarate = 3-(imidazol-4-yl)-2-oxopropyl phosphate + L-glutamate
- TIGRFAM: Amino acid biosynthesis Histidine family histidinol-phosphate transaminase (TIGR01141; EC 2.6.1.9; HMM-score: 198.5)Biosynthesis of cofactors, prosthetic groups, and carriers Heme, porphyrin, and cobalamin threonine-phosphate decarboxylase (TIGR01140; EC 4.1.1.81; HMM-score: 163.9)and 8 moreCellular processes Biosynthesis of natural products capreomycidine synthase (TIGR03947; HMM-score: 46)tyrosine/nicotianamine family aminotransferase (TIGR01265; HMM-score: 31.9)putative C-S lyase (TIGR04350; EC 4.4.-.-; HMM-score: 27.7)Energy metabolism Amino acids and amines tyrosine aminotransferase (TIGR01264; EC 2.6.1.5; HMM-score: 26.7)beta-methylarginine biosynthesis bifunctional aminotransferase (TIGR04544; EC 2.6.-.-; HMM-score: 19.9)succinyldiaminopimelate transaminase (TIGR03537; EC 2.6.1.17; HMM-score: 18.3)succinyldiaminopimelate transaminase (TIGR03539; EC 2.6.1.17; HMM-score: 17.9)LL-diaminopimelate aminotransferase (TIGR03540; EC 2.6.1.83; HMM-score: 15)
- TheSEED :
- Similar to histidinol-phosphate aminotransferase
Amino Acids and Derivatives Histidine Metabolism Histidine Biosynthesis Histidinol-phosphate aminotransferase (EC 2.6.1.9)and 1 more - PFAM: PLP_aminotran (CL0061) Aminotran_1_2; Aminotransferase class I and II (PF00155; HMM-score: 133.5)
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors: pyridoxal 5'-phosphate
- effectors:
⊟Localization[edit | edit source]
- PSORTb: Cytoplasmic
- Cytoplasmic Score: 7.5
- Cytoplasmic Membrane Score: 1.15
- Cellwall Score: 0.62
- Extracellular Score: 0.73
- Internal Helices: 0
- DeepLocPro: Cytoplasmic
- Cytoplasmic Score: 0.9989
- Cytoplasmic Membrane Score: 0.0002
- Cell wall & surface Score: 0.0001
- Extracellular Score: 0.0009
- LocateP: Intracellular
- Prediction by SwissProt Classification: Cytoplasmic
- Pathway Prediction: No pathway
- Intracellular possibility: 1
- Signal peptide possibility: -1
- N-terminally Anchored Score: 1
- Predicted Cleavage Site: No CleavageSite
- SignalP: no predicted signal peptide
- SP(Sec/SPI): 0.0182
- TAT(Tat/SPI): 0.000757
- LIPO(Sec/SPII): 0.00112
- predicted transmembrane helices (TMHMM): 0
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MIYIDKNESPVTPLDEKTMTSIISATPYNLYPDAAYEQFKEAYAKFYGLSPEQIIAGNGSDELIQKLMLIMPEGPALTLNPDFFMYQAYAAQVNREIAFVDAGSDLTFDLETILTKIDEVQPSFFIMSNPHNPSGKQFDTAFLTAIADKMKALNGYFVIDEAYLDYGTAYDVELAPHILRMRTLSKAFGIAGLRLGVLISTAGTIKHIQKIEHPYPLNVFTLNIATYIFRHREETRQFLTMQRQLAEQLKQIFDTHVADKMSVFPSNANFVLTKGSAAQQLGQYVYEQGFKPRFYDEPVMKGYVRYSIATASQLKQLEEIVKEWSAKYDLSKTTKHS
⊟Experimental data[edit | edit source]
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
- MicrobesOnline: SAOUHSC_03008 < SAOUHSC_03009 < hisH < hisB < SAOUHSC_03012 < SAOUHSC_03013 < hisG < hisZ
⊟Regulation[edit | edit source]
- regulators: CodY* (repression) regulon, HisR* (repression) regulon
CodY* (TF) important in Amino acid metabolism; RegPrecise transcription unit transferred from N315 data RegPrecise HisR* (TF) important in Histidine biosynthesis; RegPrecise transcription unit transferred from N315 data RegPrecise
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: [1]
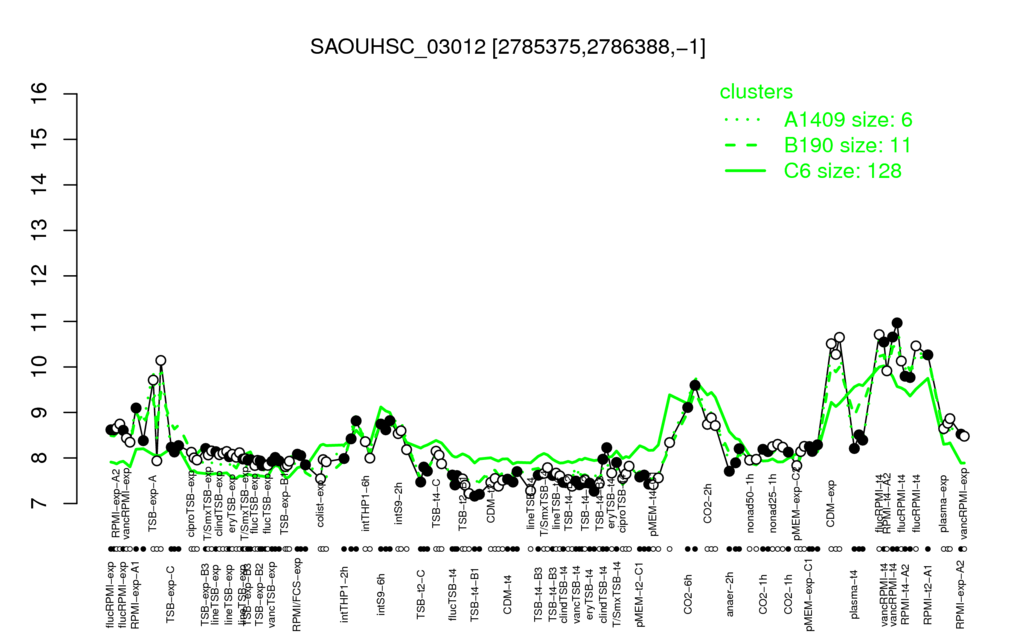 Multi-gene expression profiles
Multi-gene expression profiles
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
- Aureolib: no data available
⊟Protein stability[edit | edit source]
- half-life: no data available
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You can add further information about the gene and protein here. [edit]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ Ulrike Mäder, Pierre Nicolas, Maren Depke, Jan Pané-Farré, Michel Debarbouille, Magdalena M van der Kooi-Pol, Cyprien Guérin, Sandra Dérozier, Aurelia Hiron, Hanne Jarmer, Aurélie Leduc, Stephan Michalik, Ewoud Reilman, Marc Schaffer, Frank Schmidt, Philippe Bessières, Philippe Noirot, Michael Hecker, Tarek Msadek, Uwe Völker, Jan Maarten van Dijl
Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions.
PLoS Genet: 2016, 12(4);e1005962
[PubMed:27035918] [WorldCat.org] [DOI] (I e)
