Jump to navigation
Jump to search
NCBI: 03-AUG-2016
⊟Summary[edit | edit source]
- organism: Staphylococcus aureus NCTC8325
- locus tag: SAOUHSC_01276
- pan locus tag?: SAUPAN003612000
- symbol: glpK
- pan gene symbol?: glpK
- synonym:
- product: glycerol kinase
⊟Genome View[edit | edit source]
⊟Gene[edit | edit source]
⊟General[edit | edit source]
- type: CDS
- locus tag: SAOUHSC_01276
- symbol: glpK
- product: glycerol kinase
- replicon: chromosome
- strand: +
- coordinates: 1233281..1234777
- length: 1497
- essential: no DEG other strains
⊟Accession numbers[edit | edit source]
- Gene ID: 3919929 NCBI
- RefSeq: YP_499809 NCBI
- BioCyc: G1I0R-1194 BioCyc
- MicrobesOnline: 1289723 MicrobesOnline
⊟Phenotype[edit | edit source]
Share your knowledge and add information here. [edit]
⊟DNA sequence[edit | edit source]
- 1
61
121
181
241
301
361
421
481
541
601
661
721
781
841
901
961
1021
1081
1141
1201
1261
1321
1381
1441ATGGAAAAATATATTTTATCTATAGACCAAGGAACAACAAGCTCAAGAGCGATTTTATTC
AATCAAAAAGGGGAAATTGCAGGGGTAGCACAACGTGAGTTTAAGCAATATTTTCCACAA
TCAGGTTGGGTTGAACATGATGCAAATGAAATTTGGACATCTGTGTTAGCTGTAATGACG
GAAGTAATTAATGAAAATGATGTTAGAGCTGATCAAATTGCAGGTATCGGTATTACAAAC
CAACGTGAAACAACGGTTGTTTGGGACAAACATACTGGCCGCCCAATTTATCACGCAATT
GTTTGGCAATCACGTCAAACACAATCAATTTGTTCAGAATTAAAACAACAAGGATATGAA
CAAACATTTAGAGATAAGACAGGATTACTTTTAGATCCGTATTTTGCAGGTACAAAAGTT
AAATGGATTCTAGACAATGTTGAAGGTGCACGAGAAAAAGCAGAAAATGGCGATCTATTA
TTTGGAACGATTGATACTTGGTTAGTATGGAAATTATCAGGAAAAGCTGCGCATATTACT
GATTATTCAAATGCGAGTCGTACATTAATGTTTAATATCCATGATTTAGAATGGGACGAT
GAGTTATTAGAACTACTTACAGTACCTAAAAATATGTTGCCAGAAGTTAAAGCTTCGAGT
GAAGTATATGGTAAGACAATTGATTACCACTTCTATGGTCAAGAAGTACCAATCGCTGGA
GTAGCTGGTGATCAACAAGCAGCATTATTTGGACAAGCTTGCTTCGAACGTGGTGACGTG
AAAAACACATATGGAACTGGTGGCTTCATGTTAATGAATACAGGTGACAAAGCGGTTAAA
TCTGAAAGTGGTTTATTAACAACAATTGCTTATGGTATTGATGGAAAAGTAAATTATGCG
CTTGAAGGTTCCATCTTTGTTTCGGGTTCAGCAATCCAATGGTTACGTGATGGATTAAGA
ATGATTAATTCAGCACCACAATCAGAAAGTTATGCGACACGAGTTGACTCTACTGAGGGT
GTTTATGTTGTTCCAGCTTTTGTAGGTTTAGGAACACCATATTGGGATTCTGAAGCACGT
GGTGCGATTTTCGGTTTAACACGTGGAACTGAAAAAGAGCACTTTATCCGTGCAACTTTA
GAATCACTATGTTACCAAACTCGTGACGTTATGGAAGCAATGTCAAAAGACTCTGGTATT
GATGTCCAAAGTTTACGTGTCGATGGTGGTGCAGTTAAAAATAACTTTATTATGCAGTTC
CAAGCAGACATTGTTAATACTTCTGTTGAAAGACCTGAAATTCAAGAAACTACAGCTTTA
GGTGCTGCATTTTTGGCAGGTTTAGCAGTTGGATTCTGGGAGAGTAAAGATGATATCGCT
AAAAACTGGAAATTAGAAGAAAAATTCGATCCGAAAATGGATGAAGGCGAAAGAGAAAAA
TTATATAGAGGTTGGAAAAAAGCTGTTGAAGCAACACAAGTTTTTAAAACAGAATAA60
120
180
240
300
360
420
480
540
600
660
720
780
840
900
960
1020
1080
1140
1200
1260
1320
1380
1440
1497
⊟Protein[edit | edit source]
⊟General[edit | edit source]
- locus tag: SAOUHSC_01276
- symbol: GlpK
- description: glycerol kinase
- length: 498
- theoretical pI: 4.6683
- theoretical MW: 55625.2
- GRAVY: -0.340964
⊟Function[edit | edit source]
- reaction: EC 2.7.1.30? ExPASyGlycerol kinase ATP + glycerol = ADP + sn-glycerol 3-phosphate
- TIGRFAM: Energy metabolism Other glycerol kinase (TIGR01311; EC 2.7.1.30; HMM-score: 760.2)and 7 moreEnergy metabolism Sugars xylulokinase (TIGR01312; EC 2.7.1.17; HMM-score: 240.7)Energy metabolism Sugars gluconate kinase (TIGR01314; EC 2.7.1.12; HMM-score: 173.5)Energy metabolism Sugars L-fuculokinase (TIGR02628; EC 2.7.1.51; HMM-score: 120.9)FGGY-family pentulose kinase (TIGR01315; EC 2.7.1.-; HMM-score: 109.4)Energy metabolism Sugars ribulokinase (TIGR01234; EC 2.7.1.16; HMM-score: 68.5)rhamnulokinase (TIGR02627; EC 2.7.1.5; HMM-score: 57.7)TraF-like protein (TIGR02740; HMM-score: 12.8)
- TheSEED :
- Glycerol kinase (EC 2.7.1.30)
Carbohydrates Central carbohydrate metabolism Dihydroxyacetone kinases Glycerol kinase (EC 2.7.1.30)and 2 more - PFAM: Actin_ATPase (CL0108) FGGY_N; FGGY family of carbohydrate kinases, N-terminal domain (PF00370; HMM-score: 297)and 2 moreFGGY_C; FGGY family of carbohydrate kinases, C-terminal domain (PF02782; HMM-score: 165.5)BcrAD_BadFG; BadF/BadG/BcrA/BcrD ATPase family (PF01869; HMM-score: 19)
⊟Structure, modifications & cofactors[edit | edit source]
- domains:
- modifications:
- cofactors:
- effectors:
⊟Localization[edit | edit source]
- PSORTb: Cytoplasmic
- Cytoplasmic Score: 9.67
- Cytoplasmic Membrane Score: 0.01
- Cellwall Score: 0.15
- Extracellular Score: 0.17
- Internal Helices: 0
- DeepLocPro: Cytoplasmic
- Cytoplasmic Score: 0.9394
- Cytoplasmic Membrane Score: 0.0121
- Cell wall & surface Score: 0.0013
- Extracellular Score: 0.0472
- LocateP: Intracellular
- Prediction by SwissProt Classification: Cytoplasmic
- Pathway Prediction: No pathway
- Intracellular possibility: 1
- Signal peptide possibility: -1
- N-terminally Anchored Score: 1
- Predicted Cleavage Site: No CleavageSite
- SignalP: no predicted signal peptide
- SP(Sec/SPI): 0.004055
- TAT(Tat/SPI): 0.000391
- LIPO(Sec/SPII): 0.000446
- predicted transmembrane helices (TMHMM): 0
⊟Accession numbers[edit | edit source]
⊟Protein sequence[edit | edit source]
- MEKYILSIDQGTTSSRAILFNQKGEIAGVAQREFKQYFPQSGWVEHDANEIWTSVLAVMTEVINENDVRADQIAGIGITNQRETTVVWDKHTGRPIYHAIVWQSRQTQSICSELKQQGYEQTFRDKTGLLLDPYFAGTKVKWILDNVEGAREKAENGDLLFGTIDTWLVWKLSGKAAHITDYSNASRTLMFNIHDLEWDDELLELLTVPKNMLPEVKASSEVYGKTIDYHFYGQEVPIAGVAGDQQAALFGQACFERGDVKNTYGTGGFMLMNTGDKAVKSESGLLTTIAYGIDGKVNYALEGSIFVSGSAIQWLRDGLRMINSAPQSESYATRVDSTEGVYVVPAFVGLGTPYWDSEARGAIFGLTRGTEKEHFIRATLESLCYQTRDVMEAMSKDSGIDVQSLRVDGGAVKNNFIMQFQADIVNTSVERPEIQETTALGAAFLAGLAVGFWESKDDIAKNWKLEEKFDPKMDEGEREKLYRGWKKAVEATQVFKTE
⊟Experimental data[edit | edit source]
- experimentally validated: PeptideAtlas [1] [2]
- protein localization: data available for COL
- quantitative data / protein copy number per cell: data available for COL
- interaction partners:
SAOUHSC_01201 (acpP) acyl carrier protein [3] (data from MRSA252) SAOUHSC_02490 (adk) adenylate kinase [3] (data from MRSA252) SAOUHSC_01722 (alaS) alanyl-tRNA synthetase [3] (data from MRSA252) SAOUHSC_01471 (asnC) asparaginyl-tRNA synthetase [3] (data from MRSA252) SAOUHSC_02318 (ddl) D-alanyl-alanine synthetase A [3] (data from MRSA252) SAOUHSC_02380 (deoD) purine nucleoside phosphorylase [3] (data from MRSA252) SAOUHSC_01683 (dnaK) molecular chaperone DnaK [3] (data from MRSA252) SAOUHSC_00799 (eno) phosphopyruvate hydratase [3] (data from MRSA252) SAOUHSC_00574 (eutD) phosphotransacetylase [3] (data from MRSA252) SAOUHSC_01236 (frr) ribosome recycling factor [3] (data from MRSA252) SAOUHSC_02117 (gatA) aspartyl/glutamyl-tRNA amidotransferase subunit A [3] (data from MRSA252) SAOUHSC_02116 (gatB) aspartyl/glutamyl-tRNA amidotransferase subunit B [3] (data from MRSA252) SAOUHSC_01634 (gcvT) glycine cleavage system aminomethyltransferase T [3] (data from MRSA252) SAOUHSC_02703 (gpmA) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase [3] (data from MRSA252) SAOUHSC_02254 (groEL) chaperonin GroEL [3] (data from MRSA252) SAOUHSC_02255 (groES) co-chaperonin GroES [3] (data from MRSA252) SAOUHSC_00375 (guaA) GMP synthase [3] (data from MRSA252) SAOUHSC_01159 (ileS) isoleucyl-tRNA synthetase [3] (data from MRSA252) SAOUHSC_01246 (infB) translation initiation factor IF-2 [3] (data from MRSA252) SAOUHSC_01786 (infC) translation initiation factor IF-3 [3] (data from MRSA252) SAOUHSC_00225 (ispD) 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase [3] (data from MRSA252) SAOUHSC_01875 (leuS) leucyl-tRNA synthetase [3] (data from MRSA252) SAOUHSC_00493 (lysS) lysyl-tRNA synthetase [3] (data from MRSA252) SAOUHSC_00743 (nrdF) ribonucleotide-diphosphate reductase subunit beta [3] (data from MRSA252) SAOUHSC_01243 (nusA) transcription elongation factor NusA [3] (data from MRSA252) SAOUHSC_00900 (pgi) glucose-6-phosphate isomerase [3] (data from MRSA252) SAOUHSC_00796 (pgk) phosphoglycerate kinase [3] (data from MRSA252) SAOUHSC_00519 (rplA) 50S ribosomal protein L1 [3] (data from MRSA252) SAOUHSC_02509 (rplB) 50S ribosomal protein L2 [3] (data from MRSA252) SAOUHSC_02512 (rplC) 50S ribosomal protein L3 [3] (data from MRSA252) SAOUHSC_02511 (rplD) 50S ribosomal protein L4 [3] (data from MRSA252) SAOUHSC_02500 (rplE) 50S ribosomal protein L5 [3] (data from MRSA252) SAOUHSC_02496 (rplF) 50S ribosomal protein L6 [3] (data from MRSA252) SAOUHSC_00017 (rplI) 50S ribosomal protein L9 [3] (data from MRSA252) SAOUHSC_00520 (rplJ) 50S ribosomal protein L10 [3] (data from MRSA252) SAOUHSC_00518 (rplK) 50S ribosomal protein L11 [3] (data from MRSA252) SAOUHSC_00521 (rplL) 50S ribosomal protein L7/L12 [3] (data from MRSA252) SAOUHSC_02478 (rplM) 50S ribosomal protein L13 [3] (data from MRSA252) SAOUHSC_02502 (rplN) 50S ribosomal protein L14 [3] (data from MRSA252) SAOUHSC_02492 (rplO) 50S ribosomal protein L15 [3] (data from MRSA252) SAOUHSC_02505 (rplP) 50S ribosomal protein L16 [3] (data from MRSA252) SAOUHSC_02484 (rplQ) 50S ribosomal protein L17 [3] (data from MRSA252) SAOUHSC_02495 (rplR) 50S ribosomal protein L18 [3] (data from MRSA252) SAOUHSC_01211 (rplS) 50S ribosomal protein L19 [3] (data from MRSA252) SAOUHSC_01784 (rplT) 50S ribosomal protein L20 [3] (data from MRSA252) SAOUHSC_02507 (rplV) 50S ribosomal protein L22 [3] (data from MRSA252) SAOUHSC_02510 (rplW) 50S ribosomal protein L23 [3] (data from MRSA252) SAOUHSC_02501 (rplX) 50S ribosomal protein L24 [3] (data from MRSA252) SAOUHSC_01755 (rpmA) 50S ribosomal protein L27 [3] (data from MRSA252) SAOUHSC_01191 (rpmB) 50S ribosomal protein L28 [3] (data from MRSA252) SAOUHSC_02493 (rpmD) 50S ribosomal protein L30 [3] (data from MRSA252) SAOUHSC_02361 (rpmE2) 50S ribosomal protein L31 type B [3] (data from MRSA252) SAOUHSC_01785 (rpmI) 50S ribosomal protein L35 [3] (data from MRSA252) SAOUHSC_00524 (rpoB) DNA-directed RNA polymerase subunit beta [3] (data from MRSA252) SAOUHSC_01493 (rpsA) 30S ribosomal protein S1 [3] (data from MRSA252) SAOUHSC_01232 (rpsB) 30S ribosomal protein S2 [3] (data from MRSA252) SAOUHSC_02506 (rpsC) 30S ribosomal protein S3 [3] (data from MRSA252) SAOUHSC_01829 (rpsD) 30S ribosomal protein S4 [3] (data from MRSA252) SAOUHSC_02494 (rpsE) 30S ribosomal protein S5 [3] (data from MRSA252) SAOUHSC_00348 (rpsF) 30S ribosomal protein S6 [3] (data from MRSA252) SAOUHSC_02498 (rpsH) 30S ribosomal protein S8 [3] (data from MRSA252) SAOUHSC_02477 (rpsI) 30S ribosomal protein S9 [3] (data from MRSA252) SAOUHSC_00527 (rpsL) 30S ribosomal protein S12 [3] (data from MRSA252) SAOUHSC_02487 (rpsM) 30S ribosomal protein S13 [3] (data from MRSA252) SAOUHSC_01250 (rpsO) 30S ribosomal protein S15 [3] (data from MRSA252) SAOUHSC_01208 (rpsP) 30S ribosomal protein S16 [3] (data from MRSA252) SAOUHSC_02503 (rpsQ) 30S ribosomal protein S17 [3] (data from MRSA252) SAOUHSC_00350 (rpsR) 30S ribosomal protein S18 [3] (data from MRSA252) SAOUHSC_02508 (rpsS) 30S ribosomal protein S19 [3] (data from MRSA252) SAOUHSC_01216 (sucC) succinyl-CoA synthetase subunit beta [3] (data from MRSA252) SAOUHSC_01788 (thrS) threonyl-tRNA synthetase [3] (data from MRSA252) SAOUHSC_01779 (tig) trigger factor [3] (data from MRSA252) SAOUHSC_00797 (tpiA) triosephosphate isomerase [3] (data from MRSA252) SAOUHSC_01822 (tpx) 2-Cys peroxiredoxin [3] (data from MRSA252) SAOUHSC_01234 (tsf) elongation factor Ts [3] (data from MRSA252) SAOUHSC_02353 (upp) uracil phosphoribosyltransferase [3] (data from MRSA252) SAOUHSC_00002 DNA polymerase III subunit beta [3] (data from MRSA252) SAOUHSC_00009 seryl-tRNA synthetase [3] (data from MRSA252) SAOUHSC_00069 protein A [3] (data from MRSA252) SAOUHSC_00100 2-deoxyribose-5-phosphate aldolase [3] (data from MRSA252) SAOUHSC_00187 formate acetyltransferase [3] (data from MRSA252) SAOUHSC_00206 L-lactate dehydrogenase [3] (data from MRSA252) SAOUHSC_00284 5'-nucleotidase [3] (data from MRSA252) SAOUHSC_00336 acetyl-CoA acyltransferase [3] (data from MRSA252) SAOUHSC_00346 GTP-dependent nucleic acid-binding protein EngD [3] (data from MRSA252) SAOUHSC_00364 alkyl hydroperoxide reductase subunit F [3] (data from MRSA252) SAOUHSC_00365 alkyl hydroperoxide reductase subunit C [3] (data from MRSA252) SAOUHSC_00369 hypothetical protein [3] (data from MRSA252) SAOUHSC_00371 hypothetical protein [3] (data from MRSA252) SAOUHSC_00374 inosine-5'-monophosphate dehydrogenase [3] (data from MRSA252) SAOUHSC_00444 hypothetical protein [3] (data from MRSA252) SAOUHSC_00469 regulatory protein SpoVG [3] (data from MRSA252) SAOUHSC_00474 50S ribosomal protein L25/general stress protein Ctc [3] (data from MRSA252) SAOUHSC_00483 hypothetical protein [3] (data from MRSA252) SAOUHSC_00499 pyridoxal biosynthesis lyase PdxS [3] (data from MRSA252) SAOUHSC_00505 endopeptidase [3] (data from MRSA252) SAOUHSC_00525 DNA-directed RNA polymerase subunit beta' [3] (data from MRSA252) SAOUHSC_00528 30S ribosomal protein S7 [3] (data from MRSA252) SAOUHSC_00529 elongation factor G [3] (data from MRSA252) SAOUHSC_00530 elongation factor Tu [3] (data from MRSA252) SAOUHSC_00532 2-amino-3-ketobutyrate coenzyme A ligase [3] (data from MRSA252) SAOUHSC_00562 phosphomethylpyrimidine kinase [3] (data from MRSA252) SAOUHSC_00608 alcohol dehydrogenase [3] (data from MRSA252) SAOUHSC_00634 ABC transporter substrate-binding protein [3] (data from MRSA252) SAOUHSC_00767 hypothetical protein [3] (data from MRSA252) SAOUHSC_00795 glyceraldehyde-3-phosphate dehydrogenase [3] (data from MRSA252) SAOUHSC_00798 2,3-bisphosphoglycerate-independent phosphoglycerate mutase [3] (data from MRSA252) SAOUHSC_00834 thioredoxin [3] (data from MRSA252) SAOUHSC_00836 glycine cleavage system protein H [3] (data from MRSA252) SAOUHSC_00847 ABC transporter ATP-binding protein [3] (data from MRSA252) SAOUHSC_00878 hypothetical protein [3] (data from MRSA252) SAOUHSC_00895 glutamate dehydrogenase [3] (data from MRSA252) SAOUHSC_00906 hypothetical protein [3] (data from MRSA252) SAOUHSC_00920 3-oxoacyl-ACP synthase III [3] (data from MRSA252) SAOUHSC_00921 3-oxoacyl- synthase [3] (data from MRSA252) SAOUHSC_00937 oligoendopeptidase F [3] (data from MRSA252) SAOUHSC_00947 enoyl-(acyl carrier protein) reductase [3] (data from MRSA252) SAOUHSC_00951 hypothetical protein [3] (data from MRSA252) SAOUHSC_00985 1,4-dihydroxy-2-naphthoyl-CoA synthase [3] (data from MRSA252) SAOUHSC_01007 bifunctional 5,10-methylene-tetrahydrofolate dehydrogenase/ 5,10-methylene-tetrahydrofolate cyclohydrolase [3] (data from MRSA252) SAOUHSC_01028 phosphocarrier protein HPr [3] (data from MRSA252) SAOUHSC_01029 phosphoenolpyruvate-protein phosphotransferase [3] (data from MRSA252) SAOUHSC_01036 hypothetical protein [3] (data from MRSA252) SAOUHSC_01100 thioredoxin [3] (data from MRSA252) SAOUHSC_01149 cell division protein [3] (data from MRSA252) SAOUHSC_01150 cell division protein FtsZ [3] (data from MRSA252) SAOUHSC_01169 carbamoyl phosphate synthase small subunit [3] (data from MRSA252) SAOUHSC_01199 3-oxoacyl-(acyl-carrier-protein) reductase [3] (data from MRSA252) SAOUHSC_01218 succinyl-CoA synthetase subunit alpha [3] (data from MRSA252) SAOUHSC_01228 transcriptional repressor CodY [3] (data from MRSA252) SAOUHSC_01240 prolyl-tRNA synthetase [3] (data from MRSA252) SAOUHSC_01251 polynucleotide phosphorylase/polyadenylase [3] (data from MRSA252) SAOUHSC_01252 hypothetical protein [3] (data from MRSA252) SAOUHSC_01287 glutamine synthetase [3] (data from MRSA252) SAOUHSC_01327 catalase [3] (data from MRSA252) SAOUHSC_01337 transketolase [3] (data from MRSA252) SAOUHSC_01347 aconitate hydratase [3] (data from MRSA252) SAOUHSC_01430 PTS system transporter subunit IIA [3] (data from MRSA252) SAOUHSC_01431 methionine sulfoxide reductase B [3] (data from MRSA252) SAOUHSC_01490 DNA-binding protein HU [3] (data from MRSA252) SAOUHSC_01605 6-phosphogluconate dehydrogenase [3] (data from MRSA252) SAOUHSC_01625 elongation factor P [3] (data from MRSA252) SAOUHSC_01632 glycine dehydrogenase subunit 2 [3] (data from MRSA252) SAOUHSC_01653 superoxide dismutase [3] (data from MRSA252) SAOUHSC_01666 glycyl-tRNA synthetase [3] (data from MRSA252) SAOUHSC_01676 hypothetical protein [3] (data from MRSA252) SAOUHSC_01698 hypothetical protein [3] (data from MRSA252) SAOUHSC_01719 hypothetical protein [3] (data from MRSA252) SAOUHSC_01794 glyceraldehyde 3-phosphate dehydrogenase 2 [3] (data from MRSA252) SAOUHSC_01801 isocitrate dehydrogenase [3] (data from MRSA252) SAOUHSC_01806 pyruvate kinase [3] (data from MRSA252) SAOUHSC_01807 6-phosphofructokinase [3] (data from MRSA252) SAOUHSC_01810 NADP-dependent malic enzyme [3] (data from MRSA252) SAOUHSC_01814 hypothetical protein [3] (data from MRSA252) SAOUHSC_01819 hypothetical protein [3] (data from MRSA252) SAOUHSC_01820 acetate kinase [3] (data from MRSA252) SAOUHSC_01845 formate--tetrahydrofolate ligase [3] (data from MRSA252) SAOUHSC_01858 hypothetical protein [3] (data from MRSA252) SAOUHSC_01867 D-alanine aminotransferase [3] (data from MRSA252) SAOUHSC_01868 dipeptidase PepV [3] (data from MRSA252) SAOUHSC_01869 hypothetical protein [3] (data from MRSA252) SAOUHSC_01884 hypothetical protein [3] (data from MRSA252) SAOUHSC_01901 putative translaldolase [3] (data from MRSA252) SAOUHSC_01909 S-adenosylmethionine synthetase [3] (data from MRSA252) SAOUHSC_01910 phosphoenolpyruvate carboxykinase [3] (data from MRSA252) SAOUHSC_01968 hypothetical protein [3] (data from MRSA252) SAOUHSC_02108 ferritin [3] (data from MRSA252) SAOUHSC_02140 manganese-dependent inorganic pyrophosphatase [3] (data from MRSA252) SAOUHSC_02150 hypothetical protein [3] (data from MRSA252) SAOUHSC_02300 STAS domain-containing protein [3] (data from MRSA252) SAOUHSC_02363 aldehyde dehydrogenase [3] (data from MRSA252) SAOUHSC_02365 UDP-N-acetylglucosamine 1-carboxyvinyltransferase [3] (data from MRSA252) SAOUHSC_02366 fructose-bisphosphate aldolase [3] (data from MRSA252) SAOUHSC_02369 DNA-directed RNA polymerase subunit delta [3] (data from MRSA252) SAOUHSC_02377 pyrimidine-nucleoside phosphorylase [3] (data from MRSA252) SAOUHSC_02399 glucosamine--fructose-6-phosphate aminotransferase [3] (data from MRSA252) SAOUHSC_02425 hypothetical protein [3] (data from MRSA252) SAOUHSC_02441 alkaline shock protein 23 [3] (data from MRSA252) SAOUHSC_02485 DNA-directed RNA polymerase subunit alpha [3] (data from MRSA252) SAOUHSC_02486 30S ribosomal protein S11 [3] (data from MRSA252) SAOUHSC_02512a 30S ribosomal protein S10 [3] (data from MRSA252) SAOUHSC_02577 D-isomer specific 2-hydroxyacid dehydrogenase NAD binding domain-containing protein [3] (data from MRSA252) SAOUHSC_02830 D-lactate dehydrogenase [3] (data from MRSA252) SAOUHSC_02849 pyruvate oxidase [3] (data from MRSA252) SAOUHSC_02860 HMG-CoA synthase [3] (data from MRSA252) SAOUHSC_02869 1-pyrroline-5-carboxylate dehydrogenase [3] (data from MRSA252) SAOUHSC_02922 L-lactate dehydrogenase [3] (data from MRSA252) SAOUHSC_02926 fructose-1,6-bisphosphate aldolase [3] (data from MRSA252) SAOUHSC_02927 malate:quinone oxidoreductase [3] (data from MRSA252) SAOUHSC_02965 carbamate kinase [3] (data from MRSA252) SAOUHSC_02968 ornithine carbamoyltransferase [3] (data from MRSA252) SAOUHSC_02969 arginine deiminase [3] (data from MRSA252)
⊟Expression & Regulation[edit | edit source]
⊟Operon[edit | edit source]
⊟Regulation[edit | edit source]
- regulator: CcpA* regulon
CcpA* (TF) important in Carbon catabolism; RegPrecise transcription unit transferred from N315 data RegPrecise
⊟Transcription pattern[edit | edit source]
- S.aureus Expression Data Browser: [4]
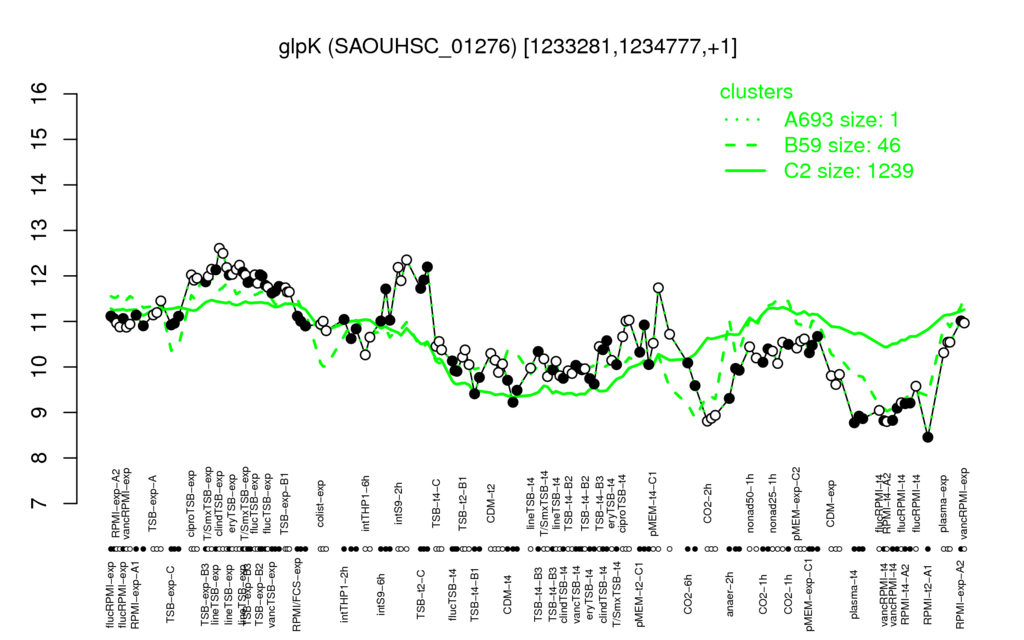 Multi-gene expression profiles
Multi-gene expression profiles
⊟Protein synthesis (provided by Aureolib)[edit | edit source]
⊟Protein stability[edit | edit source]
- half-life: no data available
⊟Biological Material[edit | edit source]
⊟Mutants[edit | edit source]
⊟Expression vector[edit | edit source]
⊟lacZ fusion[edit | edit source]
⊟GFP fusion[edit | edit source]
⊟two-hybrid system[edit | edit source]
⊟FLAG-tag construct[edit | edit source]
⊟Antibody[edit | edit source]
⊟Other Information[edit | edit source]
You can add further information about the gene and protein here. [edit]
⊟Literature[edit | edit source]
⊟References[edit | edit source]
- ↑ Maren Depke, Stephan Michalik, Alexander Rabe, Kristin Surmann, Lars Brinkmann, Nico Jehmlich, Jörg Bernhardt, Michael Hecker, Bernd Wollscheid, Zhi Sun, Robert L Moritz, Uwe Völker, Frank Schmidt
A peptide resource for the analysis of Staphylococcus aureus in host-pathogen interaction studies.
Proteomics: 2015, 15(21);3648-61
[PubMed:26224020] [WorldCat.org] [DOI] (I p) - ↑ Stephan Michalik, Maren Depke, Annette Murr, Manuela Gesell Salazar, Ulrike Kusebauch, Zhi Sun, Tanja C Meyer, Kristin Surmann, Henrike Pförtner, Petra Hildebrandt, Stefan Weiss, Laura Marcela Palma Medina, Melanie Gutjahr, Elke Hammer, Dörte Becher, Thomas Pribyl, Sven Hammerschmidt, Eric W Deutsch, Samuel L Bader, Michael Hecker, Robert L Moritz, Ulrike Mäder, Uwe Völker, Frank Schmidt
A global Staphylococcus aureus proteome resource applied to the in vivo characterization of host-pathogen interactions.
Sci Rep: 2017, 7(1);9718
[PubMed:28887440] [WorldCat.org] [DOI] (I e) - ↑ 3.000 3.001 3.002 3.003 3.004 3.005 3.006 3.007 3.008 3.009 3.010 3.011 3.012 3.013 3.014 3.015 3.016 3.017 3.018 3.019 3.020 3.021 3.022 3.023 3.024 3.025 3.026 3.027 3.028 3.029 3.030 3.031 3.032 3.033 3.034 3.035 3.036 3.037 3.038 3.039 3.040 3.041 3.042 3.043 3.044 3.045 3.046 3.047 3.048 3.049 3.050 3.051 3.052 3.053 3.054 3.055 3.056 3.057 3.058 3.059 3.060 3.061 3.062 3.063 3.064 3.065 3.066 3.067 3.068 3.069 3.070 3.071 3.072 3.073 3.074 3.075 3.076 3.077 3.078 3.079 3.080 3.081 3.082 3.083 3.084 3.085 3.086 3.087 3.088 3.089 3.090 3.091 3.092 3.093 3.094 3.095 3.096 3.097 3.098 3.099 3.100 3.101 3.102 3.103 3.104 3.105 3.106 3.107 3.108 3.109 3.110 3.111 3.112 3.113 3.114 3.115 3.116 3.117 3.118 3.119 3.120 3.121 3.122 3.123 3.124 3.125 3.126 3.127 3.128 3.129 3.130 3.131 3.132 3.133 3.134 3.135 3.136 3.137 3.138 3.139 3.140 3.141 3.142 3.143 3.144 3.145 3.146 3.147 3.148 3.149 3.150 3.151 3.152 3.153 3.154 3.155 3.156 3.157 3.158 3.159 3.160 3.161 3.162 3.163 3.164 3.165 3.166 3.167 3.168 3.169 3.170 3.171 3.172 3.173 3.174 3.175 3.176 3.177 3.178 3.179 3.180 3.181 3.182 3.183 3.184 3.185 3.186 3.187 3.188 3.189 3.190 3.191 Artem Cherkasov, Michael Hsing, Roya Zoraghi, Leonard J Foster, Raymond H See, Nikolay Stoynov, Jihong Jiang, Sukhbir Kaur, Tian Lian, Linda Jackson, Huansheng Gong, Rick Swayze, Emily Amandoron, Farhad Hormozdiari, Phuong Dao, Cenk Sahinalp, Osvaldo Santos-Filho, Peter Axerio-Cilies, Kendall Byler, William R McMaster, Robert C Brunham, B Brett Finlay, Neil E Reiner
Mapping the protein interaction network in methicillin-resistant Staphylococcus aureus.
J Proteome Res: 2011, 10(3);1139-50
[PubMed:21166474] [WorldCat.org] [DOI] (I p) - ↑ 4.0 4.1 4.2 Ulrike Mäder, Pierre Nicolas, Maren Depke, Jan Pané-Farré, Michel Debarbouille, Magdalena M van der Kooi-Pol, Cyprien Guérin, Sandra Dérozier, Aurelia Hiron, Hanne Jarmer, Aurélie Leduc, Stephan Michalik, Ewoud Reilman, Marc Schaffer, Frank Schmidt, Philippe Bessières, Philippe Noirot, Michael Hecker, Tarek Msadek, Uwe Völker, Jan Maarten van Dijl
Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions.
PLoS Genet: 2016, 12(4);e1005962
[PubMed:27035918] [WorldCat.org] [DOI] (I e)
